For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDSIIGWDKDADGIVTLTIDDLDNGANMLTDAYVEQMGAMIDRLEAEVDETTGVVITSGKSTFFAGADLT QIRSAGPEHAQQLFDGSTAIKRDLRRLETLGKPVVAAINGAALGGGLELALATHHRIAADVAGSVIGLPE VMLGVLPGAGGVARSVRLLGIQNALMQVLLTGNRFKPAKAQQVGLVDELVESVEELVPAAKTWILANPDK HVQPWDVPGFEIPGGGPATSAFAANLPAFPANLRKQLKGSRMPAPRAIMAAALEGAQVDFDTAQVIETRY FVHLVTGPIAKNMIQAFFFDLQAVQGGTSRPADVPERTINKVGILGAGMMGAGIAYVTAKAGIDVVLKDI TLQAARKGKAYSENLEQKALAKGRTTEAKSQALLDRITPTADAADFAGVDFVIEAVFENTELKNKVFAEI ENIVEPGAVLGSNTSSLPITDLAAGVSRPENFIGIHFFSPVEKMDPIEIIRGAKTSDETLARVIDYTLAI KKYPIVVNDGRGFFTTRVFSTYLLEAFNMLAEGIDPATIEQAALQAGYPAGPLQVADELSFGTIRKIMDE TVQAAAADGLPLDESVRVAAEVVTTLMDTFDRNGKQGGAGFYDYADGTRAGLWDGLRTAFNTTRALSVPF DDLQDRYLFIQAIETQKAYDDGVISSDPDANVGSIFGIGYPPWTGGVRQFATGYPGGADAFRRRADELAA AYGGRFRWI*
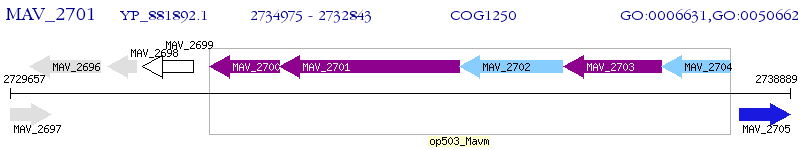
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2701 | - | - | 100% (710) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0981 | - | 0.0 | 63.57% (711) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_4681 | - | 2e-38 | 33.00% (297) | 3-hydroxybutyryl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0883 | fadB | 0.0 | 63.13% (716) | fatty oxidation protein FadB |
| M. gilvum PYR-GCK | Mflv_1692 | - | 0.0 | 62.08% (712) | 3-hydroxyacyl-CoA dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv0860 | fadB | 0.0 | 63.27% (716) | fatty oxidation protein FadB |
| M. leprae Br4923 | MLBr_02161 | fadB | 0.0 | 61.88% (711) | putative fatty oxidation complex alpha subunit |
| M. abscessus ATCC 19977 | MAB_0851 | - | 0.0 | 64.57% (714) | fatty oxidation protein FadB |
| M. marinum M | MMAR_4676 | fadB | 0.0 | 63.15% (711) | fatty oxidation protein FadB |
| M. smegmatis MC2 155 | MSMEG_5720 | - | 0.0 | 62.57% (716) | putative 3-hydroxyacyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2033 | fadB | 0.0 | 64.42% (711) | PROBABLE FATTY OXIDATION PROTEIN FADB |
| M. ulcerans Agy99 | MUL_0295 | fadB | 0.0 | 63.01% (711) | fatty oxidation protein FadB |
| M. vanbaalenii PYR-1 | Mvan_5066 | - | 0.0 | 62.46% (714) | 3-hydroxyacyl-CoA dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4676|M.marinum_M MADNTIQWDKDADGIVTLTMDDPSGSANVMNEAYIESMGKAVDRLVAEKD
MUL_0295|M.ulcerans_Agy99 MADNTIQWDKDADGIVTLTMDDPSGSANVMNEAYIESMGKAVDRLVAEKD
MLBr_02161|M.leprae_Br4923 MADTAIQWSKDADGIVTLTMNDPSRSANVMNEIYIESMGNAVDRIVAEKD
Mb0883|M.bovis_AF2122/97 MPDNTIQWDKDADGIVTLTMDDPSGSTNVMNEAYIESMGKAVDRLVAEKD
Rv0860|M.tuberculosis_H37Rv MPDNTIQWDKDADGIVTLTMDDPSGSTNVMNEAYIESMGKAVDRLVAEKD
Mflv_1692|M.gilvum_PYR-GCK MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYKASMHDAVERLVAEKD
Mvan_5066|M.vanbaalenii_PYR-1 MAENTIQWDKDADGIVTLTLDDPTGSANVMNEHYKESMHNAVEKLVAEKE
MSMEG_5720|M.smegmatis_MC2_155 MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYKESMHNAVERLAAEKD
TH_2033|M.thermoresistible__bu MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYQESMRKAVDRLEAEKD
MAB_0851|M.abscessus_ATCC_1997 MSANTIQWEKDADGIVTLTLDDPNGSANVMNDDYKQSMAAAVKRLVEEKD
MAV_2701|M.avium_104 MSDSIIGWDKDADGIVTLTIDDLDNGANMLTDAYVEQMGAMIDRLEAEVD
*. . * *.**********::* .:*::.: * .* :.:: * :
MMAR_4676|M.marinum_M SITGVVITSAKKTFFAGGDLTSMIQAGPEDAGQSFDTVETVKRQLRTLET
MUL_0295|M.ulcerans_Agy99 SITGVVITSAKKTFFAGGDLTSMIQAGPEDAGQSFDTVETVKRQLRTLET
MLBr_02161|M.leprae_Br4923 SITGVVITSAKRTFFAGGDLTAMINTGSEEAGQCFDTVETVKKQLRTLET
Mb0883|M.bovis_AF2122/97 SITGVVVASAKKTFFAGGDVKTMIQARPEDAGDVFNTVETIKRQLRTLET
Rv0860|M.tuberculosis_H37Rv SITGVVVASAKKTFFAGGDVKTMIQARPEDAGDVFNTVETIKRQLRTLET
Mflv_1692|M.gilvum_PYR-GCK SVSGVVIASAKKTFFAGGDLKGMMNVGPDNAAESFAEVEFIKADLRKLET
Mvan_5066|M.vanbaalenii_PYR-1 SITGVVIASAKKTFFAGGDLKGMMNVGPDNAAESFAEVEFIKADLRKLET
MSMEG_5720|M.smegmatis_MC2_155 SITGVVITSAKKTFFAGGDLKGMMKVGPENAAESFAEVEFIKADLRKLET
TH_2033|M.thermoresistible__bu SITGVVITSAKKTFFAGGDLNGMMTIGPDDAQQAFDMVEAVKRDLRRLET
MAB_0851|M.abscessus_ATCC_1997 SITGVVITSAKKTFFAGGDLTKIIQIQPENAQEAFNEVEEIKADLRTLET
MAV_2701|M.avium_104 ETTGVVITSGKSTFFAGADLTQIRSAGPEHAQQLFDGSTAIKRDLRRLET
. :***::*.* *****.*:. : .:.* : * :* :** ***
MMAR_4676|M.marinum_M LGKPVVAAINGAALGGGLEIALACHHRIAADAKGSQLGLPEVTLGLLPGG
MUL_0295|M.ulcerans_Agy99 LGKPVVAAINGAALGGGLEIALACHHRIAADAKGSQLGLPEATLGLLPGG
MLBr_02161|M.leprae_Br4923 LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
Mb0883|M.bovis_AF2122/97 LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
Rv0860|M.tuberculosis_H37Rv LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
Mflv_1692|M.gilvum_PYR-GCK LGVPVVAAINGAALGGGLEIALACHHRIAADTRGVVIGLPEVTLGLLPGG
Mvan_5066|M.vanbaalenii_PYR-1 LGKPVVAAINGAALGGGLEIALACHHRIAADTKGVVIGLPEVTLGLLPGG
MSMEG_5720|M.smegmatis_MC2_155 LGVPVVAAINGAALGGGLEIALACHHRIAADVKGVVIGLPEVTLGLLPGG
TH_2033|M.thermoresistible__bu LGVPVVAAINGAALGGGLEIALACHRRIAADVKGSVIGLPEVTLGLLPGG
MAB_0851|M.abscessus_ATCC_1997 FGRPVVAAINGAALGGGLEIALATHHRIAADVKGSQIGLPEVSLGLLPGG
MAV_2701|M.avium_104 LGKPVVAAINGAALGGGLELALATHHRIAADVAGSVIGLPEVMLGVLPGA
:* ****************:*** *:*****. * :****. **:***.
MMAR_4676|M.marinum_M GGVTRTVRMFGIQNAFVSILSQGTRFKPAKAKEIGLVDEVLPTVEELVPA
MUL_0295|M.ulcerans_Agy99 GGVTRTVRMFGIQNAFVSILSQGTRFKPAKAKEIGLVDEVLPTVEELVPA
MLBr_02161|M.leprae_Br4923 GGVTRTVRMFGIQNAFVNILSQGTRFKPAKAKEIGLVDELLSTVEELVPA
Mb0883|M.bovis_AF2122/97 GGVTRTVRMFGIQNAFVSVLAQGTRFKPAKAKEIGLVDELVATVEELVPA
Rv0860|M.tuberculosis_H37Rv GGVTRTVRMFGIQNAFVSVLAQGTRFKPAKAKEIGLVDELVATVEELVPA
Mflv_1692|M.gilvum_PYR-GCK GGVARTVRMFGIQKAFMEILSQGTRFKPGKAKEIGLVDELVDSVEELVPA
Mvan_5066|M.vanbaalenii_PYR-1 GGVARTVRMFGIQKAFMEILSQGTRFKPGKALEIGLVDRLVQSVDELIPA
MSMEG_5720|M.smegmatis_MC2_155 GGVARTVRMFGIQKAFMEVLSQGTRFKPGKALEVGLVDQLVSSVDELVPA
TH_2033|M.thermoresistible__bu GGVTRTVRMFGIQKAFMEILSQGTRFRPAKAKEVGLVDELVDSVDELVPR
MAB_0851|M.abscessus_ATCC_1997 GGVTRVTRMLGIQNGFVTVLAQGTRFNPTKAKEVGLIDELVGSVDELIPA
MAV_2701|M.avium_104 GGVARSVRLLGIQNALMQVLLTGNRFKPAKAQQVGLVDELVESVEELVPA
***:* .*::***:.:: :* *.**.* ** ::**:*.:: :*:**:*
MMAR_4676|M.marinum_M AKAWIK----ANPD-SHEQPWDKKGYKIPGGTPSSPGLASILPSFPALLR
MUL_0295|M.ulcerans_Agy99 AKAWIK----ANPD-SHEQPWDKKGYKIPGGTPSSPGLASILPSFPALLR
MLBr_02161|M.leprae_Br4923 AKAWIK----ANPE-AHEQPWDKKSYEMPGGTPTSPGLAAILPSFPSLLR
Mb0883|M.bovis_AF2122/97 AKAWIKEELKANPDGAGVQPWDKKGYKMPGGTPSSPGLAAILPSFPSNLR
Rv0860|M.tuberculosis_H37Rv AKAWIKEELKANPDGAGVQPWDKKGYKMPGGTPSSPGLAAILPSFPSNLR
Mflv_1692|M.gilvum_PYR-GCK AKAWI----KANPE-AHTQPWDQKGYKMPGGTPSSPGLASILPSFPALLK
Mvan_5066|M.vanbaalenii_PYR-1 AKAWI----KANPE-AHTQPWDQKGYKMPGGTPSSPGLASILPSFPALLK
MSMEG_5720|M.smegmatis_MC2_155 AKAWITEELKANPETVAVQPWDRKGYKMPGGTPSSPALASILPSFPALLR
TH_2033|M.thermoresistible__bu AKAWI----KENPD-AHTQPWDAKGYKMPGGTPSSPGLASILPSFPALLK
MAB_0851|M.abscessus_ATCC_1997 AKAWIK----ANPE--AVQRWDVKGYKIPGGTPATPALAAILPSFPSNLK
MAV_2701|M.avium_104 AKTWIL----ANPD-KHVQPWDVPGFEIPGGGPATSAFAANLPAFPANLR
**:** **: * ** .:::*** *::..:*: **:**: *:
MMAR_4676|M.marinum_M KQLKGAPMPAPKAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMI
MUL_0295|M.ulcerans_Agy99 KQLKGAPMPAPKAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMI
MLBr_02161|M.leprae_Br4923 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFVSLVTGQVAKNMI
Mb0883|M.bovis_AF2122/97 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMM
Rv0860|M.tuberculosis_H37Rv KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMM
Mflv_1692|M.gilvum_PYR-GCK KQLKGAPMPAPRAILDAAVEGAQVDFDTATRIESRYFVGLVTGQTAKNMI
Mvan_5066|M.vanbaalenii_PYR-1 KQLKGAPMPAPRAILDAAVEGAQVDFDTATRIESRYFVTLVTGQTAKNMI
MSMEG_5720|M.smegmatis_MC2_155 KQLKGAPMPAPRAILAAAVEGAQVDFDTATRIESRYFVSLVTGQTAKNMI
TH_2033|M.thermoresistible__bu KQLKGAPMPAPRAILNAAVEGAQVDFDSASRIESRYFTQLVTGQTAKNMI
MAB_0851|M.abscessus_ATCC_1997 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFTSLATGQVAKNMT
MAV_2701|M.avium_104 KQLKGSRMPAPRAIMAAALEGAQVDFDTAQVIETRYFVHLVTGPIAKNMI
*****: ****:**: **:********:* **:***. *.** ****
MMAR_4676|M.marinum_M QAFFFDLQAINAGKSRPDGIGKTPITKIGVLGAGMMGAGIAYVSAKAGYD
MUL_0295|M.ulcerans_Agy99 QAFFFDLQAINAGKSRPDGIGKTPITKIGVLGAGMMGAGIAYVSAKAGYD
MLBr_02161|M.leprae_Br4923 QAFFLDLQHINSGGSRPDGIGKTPIKKIGVLGAGMMGAGIAYVSAKAGYN
Mb0883|M.bovis_AF2122/97 QAFFFDLQAINAGGSRPEGIGKTPIKRIGVLGAGMMGAGIAYVSAKAGYE
Rv0860|M.tuberculosis_H37Rv QAFFFDLQAINAGGSRPEGIGKTPIKRIGVLGAGMMGAGIAYVSAKAGYE
Mflv_1692|M.gilvum_PYR-GCK QAFFLDLQAINGGASRPDGIAKQEIKKIGVLGAGMMGAGIAYVSAKAGYD
Mvan_5066|M.vanbaalenii_PYR-1 QAFFLDLQAINGGASRPDGIAKQEIKKIGVLGAGMMGAGIAYVSAKAGYD
MSMEG_5720|M.smegmatis_MC2_155 QAFFLDMQTINGGGSRPDGIAKQEIKKVGVLGAGMMGAGIAYVSARAGFE
TH_2033|M.thermoresistible__bu QAFFFDLQTINSGGSRPQGIEKTLPKKVGVLGAGMMGAGIAYVTAKAGFE
MAB_0851|M.abscessus_ATCC_1997 QAFFFDLQHINGGGSRPDGIAKQDIKKIGVLGAGMMGAGIAYVSAKAGYD
MAV_2701|M.avium_104 QAFFFDLQAVQGGTSRPADVPERTINKVGILGAGMMGAGIAYVTAKAGID
****:*:* ::.* *** .: : .::*:*************:*:** :
MMAR_4676|M.marinum_M VVLKDVSAEAAAKGKGYSEKLVAKALERGRTTQEKGDALLARITPTSDPA
MUL_0295|M.ulcerans_Agy99 VVLKDVSAEAAAKGKGYSEKLVAKALERGRTTQEKGDALLARITPTSDAA
MLBr_02161|M.leprae_Br4923 VVLKDVSLEAAQKGKSYSEKLEAKALERGRTTQEKSDVLLACIAPTGDPT
Mb0883|M.bovis_AF2122/97 VVLKDVSLEAAAKGKGYSEKLEAKALERGRTTQERSDALLARITPTADAA
Rv0860|M.tuberculosis_H37Rv VVLKDVSLEAAAKGKGYSEKLEAKALERGRTTQERSDALLARITPTADAA
Mflv_1692|M.gilvum_PYR-GCK VVLKDVSIEAAQKGKAYSEKIEAKALERGKTTKEKSDALLARITPAADAA
Mvan_5066|M.vanbaalenii_PYR-1 VVLKDVTLEAAQKGKAYSEKIEAKALERGRTTKEKSDALLARITPTADAA
MSMEG_5720|M.smegmatis_MC2_155 VVLKDVTLEAAERGKGYSEKIEAKALERGRTTKEKSDALLARITPTADAA
TH_2033|M.thermoresistible__bu VVLKDVTLEAAQKGKGYSEKLEAKALERGRTTPEKSKALLDRIKPTADVA
MAB_0851|M.abscessus_ATCC_1997 VVLKDVSLEAAQKGKGYSEKLEEKALSRGKTTQEKSSELLARIHPTADPA
MAV_2701|M.avium_104 VVLKDITLQAARKGKAYSENLEQKALAKGRTTEAKSQALLDRITPTADAA
*****:: :** :**.***:: *** :*:** :.. ** * *:.* :
MMAR_4676|M.marinum_M DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAVLGSNTSTLPITGLAT
MUL_0295|M.ulcerans_Agy99 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAVLGSNTSTLPITGLAT
MLBr_02161|M.leprae_Br4923 DLKGVDFVIEAVFENQELKHKVFQEIEDIVDPNAVLGSNTSTLPITGLAT
Mb0883|M.bovis_AF2122/97 DFKGVDFVIEAVFENQELKHKVFGEIEDIVEPNAILGSNTSTLPITGLAT
Rv0860|M.tuberculosis_H37Rv DFKGVDFVIEAVFENQELKHKVFGEIEDIVEPNAILGSNTSTLPITGLAT
Mflv_1692|M.gilvum_PYR-GCK DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
Mvan_5066|M.vanbaalenii_PYR-1 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
MSMEG_5720|M.smegmatis_MC2_155 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAILGSNTSTLPITGLAT
TH_2033|M.thermoresistible__bu DFKGVDFVIEAVFENTELKHKVFAEIEDIVDPKAILGSNTSTLPITGLAA
MAB_0851|M.abscessus_ATCC_1997 DLAGVDFVIEAVFENTELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
MAV_2701|M.avium_104 DFAGVDFVIEAVFENTELKNKVFAEIENIVEPGAVLGSNTSSLPITDLAA
*: ************ ***:*** ***:**:* *:******:****.**:
MMAR_4676|M.marinum_M GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MUL_0295|M.ulcerans_Agy99 GVKRQEDFIGVHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MLBr_02161|M.leprae_Br4923 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYALAIGKTP
Mb0883|M.bovis_AF2122/97 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Rv0860|M.tuberculosis_H37Rv GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Mflv_1692|M.gilvum_PYR-GCK GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Mvan_5066|M.vanbaalenii_PYR-1 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MSMEG_5720|M.smegmatis_MC2_155 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
TH_2033|M.thermoresistible__bu GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MAB_0851|M.abscessus_ATCC_1997 GVKRQEDFIGIHFFSPVDKMPLVEIIRGEKTSDEALARVFDYVLAIKKTP
MAV_2701|M.avium_104 GVSRPENFIGIHFFSPVEKMDPIEIIRGAKTSDETLARVIDYTLAIKKYP
**.* *:***:******:** :***:* *****:****:**.*** * *
MMAR_4676|M.marinum_M IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPSSIEQAGSQAGYPAPPLQ
MUL_0295|M.ulcerans_Agy99 IVVNDGRGFFASRVIGTFVNEALAMLGEGVEPSSIEQAGSQAGYPAPPLQ
MLBr_02161|M.leprae_Br4923 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPSSIEQAGAQAGYPAPPLQ
Mb0883|M.bovis_AF2122/97 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPASIEQAGSQAGYPAPPLQ
Rv0860|M.tuberculosis_H37Rv IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPASIEQAGSQAGYPAPPLQ
Mflv_1692|M.gilvum_PYR-GCK IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGAQAGYPAAPLQ
Mvan_5066|M.vanbaalenii_PYR-1 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGSQAGYPAAPLQ
MSMEG_5720|M.smegmatis_MC2_155 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGSQAGYPAPPLQ
TH_2033|M.thermoresistible__bu IVVNDSRGFYTSRVIGTFVNEALAMLGEGVEPASIEQAGGQAGYPAPPLQ
MAB_0851|M.abscessus_ATCC_1997 IVVNDSRGFFTSRVIGTFVNEAVAMLAEGIEPATIEQAGSQAGYPAPPLQ
MAV_2701|M.avium_104 IVVNDGRGFFTTRVFSTYLLEAFNMLAEGIDPATIEQAALQAGYPAGPLQ
*****.***:::**:.*:: **. **.**: .::****. ****** ***
MMAR_4676|M.marinum_M LSDELNLELMHKIATATRKGVEDAGGTYEPHP---AEAVVEKMIEVGRSG
MUL_0295|M.ulcerans_Agy99 LSDELNLELMHKIATATRKGVEDAGGTYEPHP---AEAVVEKMIEVGRSG
MLBr_02161|M.leprae_Br4923 LSDELNLELMHKIAIATRKGVEDAGGMYESHP---AETVVEKMIKLGRSG
Mb0883|M.bovis_AF2122/97 LSDELNLELMHKIAVATRKGVEDAGGTYQPHP---AEAVVEKMIELGRSG
Rv0860|M.tuberculosis_H37Rv LSDELNLELMHKIAVATRKGVEDAGGTYQPHP---AEAVVEKMIELGRSG
Mflv_1692|M.gilvum_PYR-GCK LSDELNLELMQKIATETRKATEAAGGTHVAHP---AEEVVNKMIEIGRPS
Mvan_5066|M.vanbaalenii_PYR-1 LSDELNLELMQKIANETRKATEAAGGTYEPHP---AELVVNKMIEIGRPS
MSMEG_5720|M.smegmatis_MC2_155 LSDELNLELMQKIATETRKAAEATGATYEPHP---AEAVVNKMIEIGRPS
TH_2033|M.thermoresistible__bu LSDELNLELMQKIATETRKGVEAAGGTYEPHP---AEAVVNTMIELGRPG
MAB_0851|M.abscessus_ATCC_1997 LSDELNLTLMQKIRKETAEAAKAEGKELPDDP---AGRVIDTLVEAGRPG
MAV_2701|M.avium_104 VADELSFGTIRKIMDETVQAAAADGLPLDESVRVAAEVVTTLMDTFDRNG
::***.: ::** * :.. * * * : .* .
MMAR_4676|M.marinum_M RLKGAGFYEYPEGKRAGLWPGLRETFKSGT----SEPPLQDMIDRMLFAE
MUL_0295|M.ulcerans_Agy99 RLKGAGFYEYPEGKRAGLWPGLRETFKSGT----SEPPLQDMIDRMLFAE
MLBr_02161|M.leprae_Br4923 RLKGAGFYEYVDGKRSGLWLGLRETFKSGS----SQPPLQDMIDRMLFAE
Mb0883|M.bovis_AF2122/97 RLKGAGFYEYADGKRSGLWPGLRETFKSGS----SQPPLQDMIDRMLFAE
Rv0860|M.tuberculosis_H37Rv RLKGAGFYEYADGKRSGLWPGLRETFKSGS----SQPPLQDMIDRMLFAE
Mflv_1692|M.gilvum_PYR-GCK RLKGAGFYEYVDGKRVGLWPGLADTFGSGS--GAVDIPLQDMIDRMLFAE
Mvan_5066|M.vanbaalenii_PYR-1 RLKGAGFYSYIDGKRVGLWEGLAETFGAGSGHGPATMPLQDMIDRMLFAE
MSMEG_5720|M.smegmatis_MC2_155 RLKGAGFYEYVDGKRVGLWPGLADTFGSGK----VDIPLQDMIDRMLFAE
TH_2033|M.thermoresistible__bu RAKGAGFYEYADGKRAGLWPGLREAFKSGS----STPPLQDMIDRMLFAE
MAB_0851|M.abscessus_ATCC_1997 RLGGAGFYDYVDGKRTELWPGLRETFKTRNGADPANPPLQDLIERMLFAE
MAV_2701|M.avium_104 KQGGAGFYDYADGTRAGLWDGLRTAFNTTR---ALSVPFDDLQDRYLFIQ
: *****.* :*.* ** ** :* : *::*: :* ** :
MMAR_4676|M.marinum_M ALETQKCLDENVLMSTADANIGSIMGIGFPPWTGGSAQYIVGYEGALGRG
MUL_0295|M.ulcerans_Agy99 ALETQKCLDENVLMSTADANIGSIMGIGFPPWTGGSAQYIVGYEGALGRG
MLBr_02161|M.leprae_Br4923 ALETQKCLDENVLTSTADANIGSIMGIGFPPWTGGSAQFIAGYSGPAGTG
Mb0883|M.bovis_AF2122/97 ALETQKCLDEGVLTSTADANIGSIMGIGFPPWTGGSAQFIVGYSGPAGTG
Rv0860|M.tuberculosis_H37Rv ALETQKCLDEGVLTSTADANIGSIMGIGFPPWTGGSAQFIVGYSGPAGTG
Mflv_1692|M.gilvum_PYR-GCK ALETQKCLDEGVLTSTADANIGSIMGIGFPPYTGGSAQFIVGYQGELGVG
Mvan_5066|M.vanbaalenii_PYR-1 ALETQKCLDEGVLTSTADANIGSIMGIGYPPYTGGSAQFIVGYQGELGVG
MSMEG_5720|M.smegmatis_MC2_155 ALETQKCIDEGVLTSTADANIGSIMGIGFPPYTGGSAQFIVGYQGELGVG
TH_2033|M.thermoresistible__bu ALETQKCLDEGVITSTADANIGSIMGIGFPPWTGGTAQFIVGYQGELGTG
MAB_0851|M.abscessus_ATCC_1997 AIETQKCFDEGVLTSTADANIGSIFGIGFPAWTGGVHQYILGYDGPAGKG
MAV_2701|M.avium_104 AIETQKAYDDGVISSDPDANVGSIFGIGYPPWTGGVRQFATGYPG----G
*:****. *:.*: * .***:***:***:*.:*** *: ** * *
MMAR_4676|M.marinum_M KEAFVARARELAAKYGDRFLPPDSLT--
MUL_0295|M.ulcerans_Agy99 KEAFVTRARELAAKYGDRFLPPDSLT--
MLBr_02161|M.leprae_Br4923 QQAFVSRARELVAKYGDRFLPPESLL--
Mb0883|M.bovis_AF2122/97 KAAFVARTRELAAAYGDRFLPPESLLS-
Rv0860|M.tuberculosis_H37Rv KAAFVARARELAAAYGDRFLPPESLLS-
Mflv_1692|M.gilvum_PYR-GCK KEAFVARAKQLAERYGERFNPPASLTS-
Mvan_5066|M.vanbaalenii_PYR-1 KDAFVARAKELAAKYGERFNPPASLTS-
MSMEG_5720|M.smegmatis_MC2_155 KEAFVARAKQLAERYGERFNPPASLES-
TH_2033|M.thermoresistible__bu KEAFVARAKQLAERYGDRFNPPESLL--
MAB_0851|M.abscessus_ATCC_1997 KAGFVARAKELAAKYGDRFNPPASLLDA
MAV_2701|M.avium_104 ADAFRRRADELAAAYGGRFRWI------
.* *: :*. ** **