For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSANTIQWEKDADGIVTLTLDDPNGSANVMNDDYKQSMAAAVKRLVEEKDSITGVVITSAKKTFFAGGDL TKIIQIQPENAQEAFNEVEEIKADLRTLETFGRPVVAAINGAALGGGLEIALATHHRIAADVKGSQIGLP EVSLGLLPGGGGVTRVTRMLGIQNGFVTVLAQGTRFNPTKAKEVGLIDELVGSVDELIPAAKAWIKANPE AVQRWDVKGYKIPGGTPATPALAAILPSFPSNLKKQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRY FTSLATGQVAKNMTQAFFFDLQHINGGGSRPDGIAKQDIKKIGVLGAGMMGAGIAYVSAKAGYDVVLKDV SLEAAQKGKGYSEKLEEKALSRGKTTQEKSSELLARIHPTADPADLAGVDFVIEAVFENTELKHKVFQEI EDIVEPNALLGSNTSTLPITGLATGVKRQEDFIGIHFFSPVDKMPLVEIIRGEKTSDEALARVFDYVLAI KKTPIVVNDSRGFFTSRVIGTFVNEAVAMLAEGIEPATIEQAGSQAGYPAPPLQLSDELNLTLMQKIRKE TAEAAKAEGKELPDDPAGRVIDTLVEAGRPGRLGGAGFYDYVDGKRTELWPGLRETFKTRNGADPANPPL QDLIERMLFAEAIETQKCFDEGVLTSTADANIGSIFGIGFPAWTGGVHQYILGYDGPAGKGKAGFVARAK ELAAKYGDRFNPPASLLDA
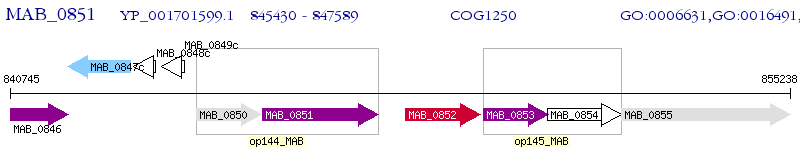
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0851 | - | - | 100% (719) | fatty oxidation protein FadB |
| M. abscessus ATCC 19977 | MAB_4094c | - | 6e-39 | 34.95% (289) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0904 | - | 5e-32 | 33.10% (281) | putative 3-hydroxyacyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0883 | fadB | 0.0 | 78.70% (723) | fatty oxidation protein FadB |
| M. gilvum PYR-GCK | Mflv_1692 | - | 0.0 | 77.96% (717) | 3-hydroxyacyl-CoA dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv0860 | fadB | 0.0 | 78.84% (723) | fatty oxidation protein FadB |
| M. leprae Br4923 | MLBr_02161 | fadB | 0.0 | 77.30% (718) | putative fatty oxidation complex alpha subunit |
| M. marinum M | MMAR_4676 | fadB | 0.0 | 77.96% (717) | fatty oxidation protein FadB |
| M. avium 104 | MAV_0981 | - | 0.0 | 79.08% (717) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5720 | - | 0.0 | 78.12% (722) | putative 3-hydroxyacyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2033 | fadB | 0.0 | 77.02% (718) | PROBABLE FATTY OXIDATION PROTEIN FADB |
| M. ulcerans Agy99 | MUL_0295 | fadB | 0.0 | 77.13% (717) | fatty oxidation protein FadB |
| M. vanbaalenii PYR-1 | Mvan_5066 | - | 0.0 | 78.66% (717) | 3-hydroxyacyl-CoA dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1692|M.gilvum_PYR-GCK MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYKASMHDAVERLVAEKD
Mvan_5066|M.vanbaalenii_PYR-1 MAENTIQWDKDADGIVTLTLDDPTGSANVMNEHYKESMHNAVEKLVAEKE
MSMEG_5720|M.smegmatis_MC2_155 MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYKESMHNAVERLAAEKD
TH_2033|M.thermoresistible__bu MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYQESMRKAVDRLEAEKD
MAV_0981|M.avium_104 MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYKESMHNAVERLVAEKD
Mb0883|M.bovis_AF2122/97 MPDNTIQWDKDADGIVTLTMDDPSGSTNVMNEAYIESMGKAVDRLVAEKD
Rv0860|M.tuberculosis_H37Rv MPDNTIQWDKDADGIVTLTMDDPSGSTNVMNEAYIESMGKAVDRLVAEKD
MMAR_4676|M.marinum_M MADNTIQWDKDADGIVTLTMDDPSGSANVMNEAYIESMGKAVDRLVAEKD
MUL_0295|M.ulcerans_Agy99 MADNTIQWDKDADGIVTLTMDDPSGSANVMNEAYIESMGKAVDRLVAEKD
MLBr_02161|M.leprae_Br4923 MADTAIQWSKDADGIVTLTMNDPSRSANVMNEIYIESMGNAVDRIVAEKD
MAB_0851|M.abscessus_ATCC_1997 MSANTIQWEKDADGIVTLTLDDPNGSANVMNDDYKQSMAAAVKRLVEEKD
*. .:*:*.**********::**. *:****: * ** **.:: **:
Mflv_1692|M.gilvum_PYR-GCK SVSGVVIASAKKTFFAGGDLKGMMNVGPDNAAESFAEVEFIKADLRKLET
Mvan_5066|M.vanbaalenii_PYR-1 SITGVVIASAKKTFFAGGDLKGMMNVGPDNAAESFAEVEFIKADLRKLET
MSMEG_5720|M.smegmatis_MC2_155 SITGVVITSAKKTFFAGGDLKGMMKVGPENAAESFAEVEFIKADLRKLET
TH_2033|M.thermoresistible__bu SITGVVITSAKKTFFAGGDLNGMMTIGPDDAQQAFDMVEAVKRDLRRLET
MAV_0981|M.avium_104 SITGVVITSAKKTFFAGGDLKSMINLGPENAGEAFDTVEAVKRDLRALET
Mb0883|M.bovis_AF2122/97 SITGVVVASAKKTFFAGGDVKTMIQARPEDAGDVFNTVETIKRQLRTLET
Rv0860|M.tuberculosis_H37Rv SITGVVVASAKKTFFAGGDVKTMIQARPEDAGDVFNTVETIKRQLRTLET
MMAR_4676|M.marinum_M SITGVVITSAKKTFFAGGDLTSMIQAGPEDAGQSFDTVETVKRQLRTLET
MUL_0295|M.ulcerans_Agy99 SITGVVITSAKKTFFAGGDLTSMIQAGPEDAGQSFDTVETVKRQLRTLET
MLBr_02161|M.leprae_Br4923 SITGVVITSAKRTFFAGGDLTAMINTGSEEAGQCFDTVETVKKQLRTLET
MAB_0851|M.abscessus_ATCC_1997 SITGVVITSAKKTFFAGGDLTKIIQIQPENAQEAFNEVEEIKADLRTLET
*::***::***:*******:. :: .::* : * ** :* :** ***
Mflv_1692|M.gilvum_PYR-GCK LGVPVVAAINGAALGGGLEIALACHHRIAADTRGVVIGLPEVTLGLLPGG
Mvan_5066|M.vanbaalenii_PYR-1 LGKPVVAAINGAALGGGLEIALACHHRIAADTKGVVIGLPEVTLGLLPGG
MSMEG_5720|M.smegmatis_MC2_155 LGVPVVAAINGAALGGGLEIALACHHRIAADVKGVVIGLPEVTLGLLPGG
TH_2033|M.thermoresistible__bu LGVPVVAAINGAALGGGLEIALACHRRIAADVKGSVIGLPEVTLGLLPGG
MAV_0981|M.avium_104 LGKPVVAAINGAALGGGLEIALACHHRIAADVKGLVVGLPEVTLGLLPGG
Mb0883|M.bovis_AF2122/97 LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
Rv0860|M.tuberculosis_H37Rv LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
MMAR_4676|M.marinum_M LGKPVVAAINGAALGGGLEIALACHHRIAADAKGSQLGLPEVTLGLLPGG
MUL_0295|M.ulcerans_Agy99 LGKPVVAAINGAALGGGLEIALACHHRIAADAKGSQLGLPEATLGLLPGG
MLBr_02161|M.leprae_Br4923 LGKPVVAAINGAALGGGLEIALACHHRIAADVKGSQLGLPEVTLGLLPGG
MAB_0851|M.abscessus_ATCC_1997 FGRPVVAAINGAALGGGLEIALATHHRIAADVKGSQIGLPEVSLGLLPGG
:* ******************** *:*****.:* :****.:*******
Mflv_1692|M.gilvum_PYR-GCK GGVARTVRMFGIQKAFMEILSQGTRFKPGKAKEIGLVDELVDSVEELVPA
Mvan_5066|M.vanbaalenii_PYR-1 GGVARTVRMFGIQKAFMEILSQGTRFKPGKALEIGLVDRLVQSVDELIPA
MSMEG_5720|M.smegmatis_MC2_155 GGVARTVRMFGIQKAFMEVLSQGTRFKPGKALEVGLVDQLVSSVDELVPA
TH_2033|M.thermoresistible__bu GGVTRTVRMFGIQKAFMEILSQGTRFRPAKAKEVGLVDELVDSVDELVPR
MAV_0981|M.avium_104 GGVTRTVRMFGIQNAFMNILSQGTRFKPDKAKEIGLVDELVGSVEELVPA
Mb0883|M.bovis_AF2122/97 GGVTRTVRMFGIQNAFVSVLAQGTRFKPAKAKEIGLVDELVATVEELVPA
Rv0860|M.tuberculosis_H37Rv GGVTRTVRMFGIQNAFVSVLAQGTRFKPAKAKEIGLVDELVATVEELVPA
MMAR_4676|M.marinum_M GGVTRTVRMFGIQNAFVSILSQGTRFKPAKAKEIGLVDEVLPTVEELVPA
MUL_0295|M.ulcerans_Agy99 GGVTRTVRMFGIQNAFVSILSQGTRFKPAKAKEIGLVDEVLPTVEELVPA
MLBr_02161|M.leprae_Br4923 GGVTRTVRMFGIQNAFVNILSQGTRFKPAKAKEIGLVDELLSTVEELVPA
MAB_0851|M.abscessus_ATCC_1997 GGVTRVTRMLGIQNGFVTVLAQGTRFNPTKAKEVGLIDELVGSVDELIPA
***:*..**:***:.*: :*:*****.* ** *:**:*.:: :*:**:*
Mflv_1692|M.gilvum_PYR-GCK AKAWI----KANPE-AHTQPWDQKGYKMPGGTPSSPGLASILPSFPALLK
Mvan_5066|M.vanbaalenii_PYR-1 AKAWI----KANPE-AHTQPWDQKGYKMPGGTPSSPGLASILPSFPALLK
MSMEG_5720|M.smegmatis_MC2_155 AKAWITEELKANPETVAVQPWDRKGYKMPGGTPSSPALASILPSFPALLR
TH_2033|M.thermoresistible__bu AKAWI----KENPD-AHTQPWDAKGYKMPGGTPSSPGLASILPSFPALLK
MAV_0981|M.avium_104 AKAWI----KANPD-AHEQPWDKKGYKMPGGTPSSPALAGILPSFPALLK
Mb0883|M.bovis_AF2122/97 AKAWIKEELKANPDGAGVQPWDKKGYKMPGGTPSSPGLAAILPSFPSNLR
Rv0860|M.tuberculosis_H37Rv AKAWIKEELKANPDGAGVQPWDKKGYKMPGGTPSSPGLAAILPSFPSNLR
MMAR_4676|M.marinum_M AKAWIK----ANPD-SHEQPWDKKGYKIPGGTPSSPGLASILPSFPALLR
MUL_0295|M.ulcerans_Agy99 AKAWIK----ANPD-SHEQPWDKKGYKIPGGTPSSPGLASILPSFPALLR
MLBr_02161|M.leprae_Br4923 AKAWIK----ANPE-AHEQPWDKKSYEMPGGTPTSPGLAAILPSFPSLLR
MAB_0851|M.abscessus_ATCC_1997 AKAWIK----ANPE--AVQRWDVKGYKIPGGTPATPALAAILPSFPSNLK
***** **: * ** *.*::*****::*.**.******: *:
Mflv_1692|M.gilvum_PYR-GCK KQLKGAPMPAPRAILDAAVEGAQVDFDTATRIESRYFVGLVTGQTAKNMI
Mvan_5066|M.vanbaalenii_PYR-1 KQLKGAPMPAPRAILDAAVEGAQVDFDTATRIESRYFVTLVTGQTAKNMI
MSMEG_5720|M.smegmatis_MC2_155 KQLKGAPMPAPRAILAAAVEGAQVDFDTATRIESRYFVSLVTGQTAKNMI
TH_2033|M.thermoresistible__bu KQLKGAPMPAPRAILNAAVEGAQVDFDSASRIESRYFTQLVTGQTAKNMI
MAV_0981|M.avium_104 KQLKGAPMPAPRAILDAAVEGAQVDFDTASRIESRYFTQLVTGQVAKNMI
Mb0883|M.bovis_AF2122/97 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMM
Rv0860|M.tuberculosis_H37Rv KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMM
MMAR_4676|M.marinum_M KQLKGAPMPAPKAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMI
MUL_0295|M.ulcerans_Agy99 KQLKGAPMPAPKAILAAAVEGAQVDFDTASRIESRYFASLVTGQVAKNMI
MLBr_02161|M.leprae_Br4923 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFVSLVTGQVAKNMI
MAB_0851|M.abscessus_ATCC_1997 KQLKGAPMPAPRAILAAAVEGAQVDFDTASRIESRYFTSLATGQVAKNMT
***********:*** ***********:*:*******. *.***.****
Mflv_1692|M.gilvum_PYR-GCK QAFFLDLQAINGGASRPDGIAKQEIKKIGVLGAGMMGAGIAYVSAKAGYD
Mvan_5066|M.vanbaalenii_PYR-1 QAFFLDLQAINGGASRPDGIAKQEIKKIGVLGAGMMGAGIAYVSAKAGYD
MSMEG_5720|M.smegmatis_MC2_155 QAFFLDMQTINGGGSRPDGIAKQEIKKVGVLGAGMMGAGIAYVSARAGFE
TH_2033|M.thermoresistible__bu QAFFFDLQTINSGGSRPQGIEKTLPKKVGVLGAGMMGAGIAYVTAKAGFE
MAV_0981|M.avium_104 QAFFFDLQHINGGGSRPEGIEPVKINKIGVLGAGMMGAGIAYVSAKAGFD
Mb0883|M.bovis_AF2122/97 QAFFFDLQAINAGGSRPEGIGKTPIKRIGVLGAGMMGAGIAYVSAKAGYE
Rv0860|M.tuberculosis_H37Rv QAFFFDLQAINAGGSRPEGIGKTPIKRIGVLGAGMMGAGIAYVSAKAGYE
MMAR_4676|M.marinum_M QAFFFDLQAINAGKSRPDGIGKTPITKIGVLGAGMMGAGIAYVSAKAGYD
MUL_0295|M.ulcerans_Agy99 QAFFFDLQAINAGKSRPDGIGKTPITKIGVLGAGMMGAGIAYVSAKAGYD
MLBr_02161|M.leprae_Br4923 QAFFLDLQHINSGGSRPDGIGKTPIKKIGVLGAGMMGAGIAYVSAKAGYN
MAB_0851|M.abscessus_ATCC_1997 QAFFFDLQHINGGGSRPDGIAKQDIKKIGVLGAGMMGAGIAYVSAKAGYD
****:*:* **.* ***:** .::***************:*:**::
Mflv_1692|M.gilvum_PYR-GCK VVLKDVSIEAAQKGKAYSEKIEAKALERGKTTKEKSDALLARITPAADAA
Mvan_5066|M.vanbaalenii_PYR-1 VVLKDVTLEAAQKGKAYSEKIEAKALERGRTTKEKSDALLARITPTADAA
MSMEG_5720|M.smegmatis_MC2_155 VVLKDVTLEAAERGKGYSEKIEAKALERGRTTKEKSDALLARITPTADAA
TH_2033|M.thermoresistible__bu VVLKDVTLEAAQKGKGYSEKLEAKALERGRTTPEKSKALLDRIKPTADVA
MAV_0981|M.avium_104 VVLKDVSLEAAQKGKGYSEKLEAKALQRGKTTEEKSKSLLDRITPTADAA
Mb0883|M.bovis_AF2122/97 VVLKDVSLEAAAKGKGYSEKLEAKALERGRTTQERSDALLARITPTADAA
Rv0860|M.tuberculosis_H37Rv VVLKDVSLEAAAKGKGYSEKLEAKALERGRTTQERSDALLARITPTADAA
MMAR_4676|M.marinum_M VVLKDVSAEAAAKGKGYSEKLVAKALERGRTTQEKGDALLARITPTSDPA
MUL_0295|M.ulcerans_Agy99 VVLKDVSAEAAAKGKGYSEKLVAKALERGRTTQEKGDALLARITPTSDAA
MLBr_02161|M.leprae_Br4923 VVLKDVSLEAAQKGKSYSEKLEAKALERGRTTQEKSDVLLACIAPTGDPT
MAB_0851|M.abscessus_ATCC_1997 VVLKDVSLEAAQKGKGYSEKLEEKALSRGKTTQEKSSELLARIHPTADPA
******: *** :**.****: ***.**:** *:.. ** * *:.* :
Mflv_1692|M.gilvum_PYR-GCK DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
Mvan_5066|M.vanbaalenii_PYR-1 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
MSMEG_5720|M.smegmatis_MC2_155 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAILGSNTSTLPITGLAT
TH_2033|M.thermoresistible__bu DFKGVDFVIEAVFENTELKHKVFAEIEDIVDPKAILGSNTSTLPITGLAA
MAV_0981|M.avium_104 DFKGVDFVIEAVFENQELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
Mb0883|M.bovis_AF2122/97 DFKGVDFVIEAVFENQELKHKVFGEIEDIVEPNAILGSNTSTLPITGLAT
Rv0860|M.tuberculosis_H37Rv DFKGVDFVIEAVFENQELKHKVFGEIEDIVEPNAILGSNTSTLPITGLAT
MMAR_4676|M.marinum_M DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAVLGSNTSTLPITGLAT
MUL_0295|M.ulcerans_Agy99 DLKGVDFVIEAVFENQELKHKVFQEIEDIVEPNAVLGSNTSTLPITGLAT
MLBr_02161|M.leprae_Br4923 DLKGVDFVIEAVFENQELKHKVFQEIEDIVDPNAVLGSNTSTLPITGLAT
MAB_0851|M.abscessus_ATCC_1997 DLAGVDFVIEAVFENTELKHKVFQEIEDIVEPNALLGSNTSTLPITGLAT
*: ************ ******* ******:*:*:**************:
Mflv_1692|M.gilvum_PYR-GCK GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Mvan_5066|M.vanbaalenii_PYR-1 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MSMEG_5720|M.smegmatis_MC2_155 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
TH_2033|M.thermoresistible__bu GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MAV_0981|M.avium_104 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Mb0883|M.bovis_AF2122/97 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
Rv0860|M.tuberculosis_H37Rv GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MMAR_4676|M.marinum_M GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MUL_0295|M.ulcerans_Agy99 GVKRQEDFIGVHFFSPVDKMPLVEIIKGEKTSDEALARVFDYTLAIGKTP
MLBr_02161|M.leprae_Br4923 GVKRQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYALAIGKTP
MAB_0851|M.abscessus_ATCC_1997 GVKRQEDFIGIHFFSPVDKMPLVEIIRGEKTSDEALARVFDYVLAIKKTP
**********:***************:***************.*** ***
Mflv_1692|M.gilvum_PYR-GCK IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGAQAGYPAAPLQ
Mvan_5066|M.vanbaalenii_PYR-1 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGSQAGYPAAPLQ
MSMEG_5720|M.smegmatis_MC2_155 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVPAASIEQAGSQAGYPAPPLQ
TH_2033|M.thermoresistible__bu IVVNDSRGFYTSRVIGTFVNEALAMLGEGVEPASIEQAGGQAGYPAPPLQ
MAV_0981|M.avium_104 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVAPASIEHAGSQAGYPSPPLQ
Mb0883|M.bovis_AF2122/97 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPASIEQAGSQAGYPAPPLQ
Rv0860|M.tuberculosis_H37Rv IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPASIEQAGSQAGYPAPPLQ
MMAR_4676|M.marinum_M IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPSSIEQAGSQAGYPAPPLQ
MUL_0295|M.ulcerans_Agy99 IVVNDGRGFFASRVIGTFVNEALAMLGEGVEPSSIEQAGSQAGYPAPPLQ
MLBr_02161|M.leprae_Br4923 IVVNDSRGFFTSRVIGTFVNEALAMLGEGVEPSSIEQAGAQAGYPAPPLQ
MAB_0851|M.abscessus_ATCC_1997 IVVNDSRGFFTSRVIGTFVNEAVAMLAEGIEPATIEQAGSQAGYPAPPLQ
*****.***::***********:***.**: .::**:**.*****:.***
Mflv_1692|M.gilvum_PYR-GCK LSDELNLELMQKIATETRKATEAAGGTHVAHPAEEVVNKMIEIGRPSRLK
Mvan_5066|M.vanbaalenii_PYR-1 LSDELNLELMQKIANETRKATEAAGGTYEPHPAELVVNKMIEIGRPSRLK
MSMEG_5720|M.smegmatis_MC2_155 LSDELNLELMQKIATETRKAAEATGATYEPHPAEAVVNKMIEIGRPSRLK
TH_2033|M.thermoresistible__bu LSDELNLELMQKIATETRKGVEAAGGTYEPHPAEAVVNTMIELGRPGRAK
MAV_0981|M.avium_104 LSDELNLELMHKIAVATRKGIEDAGGTYEPHPAEAVVEKMIEIGRPSRLK
Mb0883|M.bovis_AF2122/97 LSDELNLELMHKIAVATRKGVEDAGGTYQPHPAEAVVEKMIELGRSGRLK
Rv0860|M.tuberculosis_H37Rv LSDELNLELMHKIAVATRKGVEDAGGTYQPHPAEAVVEKMIELGRSGRLK
MMAR_4676|M.marinum_M LSDELNLELMHKIATATRKGVEDAGGTYEPHPAEAVVEKMIEVGRSGRLK
MUL_0295|M.ulcerans_Agy99 LSDELNLELMHKIATATRKGVEDAGGTYEPHPAEAVVEKMIEVGRSGRLK
MLBr_02161|M.leprae_Br4923 LSDELNLELMHKIAIATRKGVEDAGGMYESHPAETVVEKMIKLGRSGRLK
MAB_0851|M.abscessus_ATCC_1997 LSDELNLTLMQKIRKETAEAAKAEGKELPDDPAGRVIDTLVEAGRPGRLG
******* **:** * :. : * .** *::.::: **..*
Mflv_1692|M.gilvum_PYR-GCK GAGFYEYVDGKRVGLWPGLADTFGSGS--GAVDIPLQDMIDRMLFAEALE
Mvan_5066|M.vanbaalenii_PYR-1 GAGFYSYIDGKRVGLWEGLAETFGAGSGHGPATMPLQDMIDRMLFAEALE
MSMEG_5720|M.smegmatis_MC2_155 GAGFYEYVDGKRVGLWPGLADTFGSGK----VDIPLQDMIDRMLFAEALE
TH_2033|M.thermoresistible__bu GAGFYEYADGKRAGLWPGLREAFKSGS----STPPLQDMIDRMLFAEALE
MAV_0981|M.avium_104 GAGFYEYVDGKRTRLWPGLQETFNSGS----TSIPLQDMIDRMLFAEALE
Mb0883|M.bovis_AF2122/97 GAGFYEYADGKRSGLWPGLRETFKSGS----SQPPLQDMIDRMLFAEALE
Rv0860|M.tuberculosis_H37Rv GAGFYEYADGKRSGLWPGLRETFKSGS----SQPPLQDMIDRMLFAEALE
MMAR_4676|M.marinum_M GAGFYEYPEGKRAGLWPGLRETFKSGT----SEPPLQDMIDRMLFAEALE
MUL_0295|M.ulcerans_Agy99 GAGFYEYPEGKRAGLWPGLRETFKSGT----SEPPLQDMIDRMLFAEALE
MLBr_02161|M.leprae_Br4923 GAGFYEYVDGKRSGLWLGLRETFKSGS----SQPPLQDMIDRMLFAEALE
MAB_0851|M.abscessus_ATCC_1997 GAGFYDYVDGKRTELWPGLRETFKTRNGADPANPPLQDLIERMLFAEAIE
*****.* :*** ** ** ::* : . ****:*:*******:*
Mflv_1692|M.gilvum_PYR-GCK TQKCLDEGVLTSTADANIGSIMGIGFPPYTGGSAQFIVGYQGELGVGKEA
Mvan_5066|M.vanbaalenii_PYR-1 TQKCLDEGVLTSTADANIGSIMGIGYPPYTGGSAQFIVGYQGELGVGKDA
MSMEG_5720|M.smegmatis_MC2_155 TQKCIDEGVLTSTADANIGSIMGIGFPPYTGGSAQFIVGYQGELGVGKEA
TH_2033|M.thermoresistible__bu TQKCLDEGVITSTADANIGSIMGIGFPPWTGGTAQFIVGYQGELGTGKEA
MAV_0981|M.avium_104 TQKCLDEGVLTSTADANIGSIMGIGFPPYTGGSAQFIVGYSGAGGTGKEA
Mb0883|M.bovis_AF2122/97 TQKCLDEGVLTSTADANIGSIMGIGFPPWTGGSAQFIVGYSGPAGTGKAA
Rv0860|M.tuberculosis_H37Rv TQKCLDEGVLTSTADANIGSIMGIGFPPWTGGSAQFIVGYSGPAGTGKAA
MMAR_4676|M.marinum_M TQKCLDENVLMSTADANIGSIMGIGFPPWTGGSAQYIVGYEGALGRGKEA
MUL_0295|M.ulcerans_Agy99 TQKCLDENVLMSTADANIGSIMGIGFPPWTGGSAQYIVGYEGALGRGKEA
MLBr_02161|M.leprae_Br4923 TQKCLDENVLTSTADANIGSIMGIGFPPWTGGSAQFIAGYSGPAGTGQQA
MAB_0851|M.abscessus_ATCC_1997 TQKCFDEGVLTSTADANIGSIFGIGFPAWTGGVHQYILGYDGPAGKGKAG
****:**.*: **********:***:*.:*** *:* **.* * *: .
Mflv_1692|M.gilvum_PYR-GCK FVARAKQLAERYGERFNPPASLTS-
Mvan_5066|M.vanbaalenii_PYR-1 FVARAKELAAKYGERFNPPASLTS-
MSMEG_5720|M.smegmatis_MC2_155 FVARAKQLAERYGERFNPPASLES-
TH_2033|M.thermoresistible__bu FVARAKQLAERYGDRFNPPESLL--
MAV_0981|M.avium_104 FVARAKELAARYGDRFLPPASLTS-
Mb0883|M.bovis_AF2122/97 FVARTRELAAAYGDRFLPPESLLS-
Rv0860|M.tuberculosis_H37Rv FVARARELAAAYGDRFLPPESLLS-
MMAR_4676|M.marinum_M FVARARELAAKYGDRFLPPDSLT--
MUL_0295|M.ulcerans_Agy99 FVTRARELAAKYGDRFLPPDSLT--
MLBr_02161|M.leprae_Br4923 FVSRARELVAKYGDRFLPPESLL--
MAB_0851|M.abscessus_ATCC_1997 FVARAKELAAKYGDRFNPPASLLDA
**:*:::*. **:** ** **