For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THSARFAPKALLFDVQGTVTDFHSTVCSEAQRICGSRNQNVDWSDFVNSWRAAYFSCLEAAKPDRDNWIT VHSVYRSSLESLLAEYSITDLTAAERDELTLAWQRLVPWPDVLPGLTRLKKKFIIATLSNADVSALINIA KRGGLPWDAVFAAEMAGVFKPDPAIYHMAARYLGLAPRQIMMVASHKYDIRAAASLGFQTAFVTRPFEFG PLGLADTAYDDAFDVNAADLLDLATQLSC*
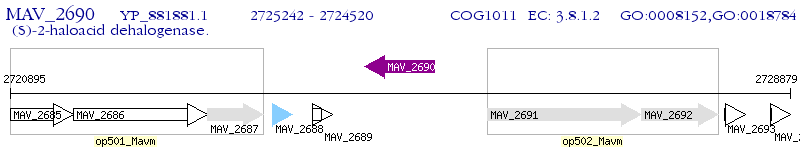
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2690 | dehII | - | 100% (240) | haloacid dehalogenase, type II |
| M. avium 104 | MAV_1053 | - | 5e-06 | 29.82% (114) | hypothetical protein MAV_1053 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0739 | - | 4e-49 | 42.79% (229) | haloacid dehalogenase, type II |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2863 | - | 7e-48 | 40.71% (226) | HAD-superfamily hydrolase YfnB |
| M. marinum M | MMAR_3371 | - | 2e-87 | 66.09% (230) | hypothetical protein MMAR_3371 |
| M. smegmatis MC2 155 | MSMEG_3450 | dehII | 1e-35 | 37.66% (231) | haloacid dehalogenase, type II |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1268 | - | 4e-77 | 66.50% (200) | hypothetical protein MUL_1268 |
| M. vanbaalenii PYR-1 | Mvan_0108 | - | 5e-48 | 44.10% (229) | haloacid dehalogenase, type II |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3371|M.marinum_M ----MSLMVTALLFDVQGTATDFHSTVCDEARRISAGRHPDADWPELVRR
MUL_1268|M.ulcerans_Agy99 -----------MLFDVQGTATDFHSTVCDEARRISAGRHPDADWPELVRR
MAV_2690|M.avium_104 MTHSARFAPKALLFDVQGTVTDFHSTVCSEAQRICGSRNQNVDWSDFVNS
MAB_2863|M.abscessus_ATCC_1997 ------MRYKAYLFDVQGTLLDFFEPVS----RAVAEYSPDVDAAAFTRA
Mflv_0739|M.gilvum_PYR-GCK ------MPIRALAFDVFGTVVDWRSSIIAELSEFGREHGVDADWSRFADD
Mvan_0108|M.vanbaalenii_PYR-1 MREEQTVVIRALAFDVFGTVVDWRSSVIRELGDFGRRNGVSADWPRFADD
MSMEG_3450|M.smegmatis_MC2_155 ------MALRALVFDVFGTLVDWRSGVADAF----RSAGVVGDPDDLADA
*** ** *: . : * :.
MMAR_3371|M.marinum_M WRAGYFSALDASSGGHGGWVSVYSVYRQVLDAVLDGCGVTGLSAAERDEL
MUL_1268|M.ulcerans_Agy99 WRAGYFSALDASSGGHGGWVSVYSVYRQVLDAVLDGCGVTGLSAAERDEL
MAV_2690|M.avium_104 WRAAYFSCLEAAKPDRDNWITVHSVYRSSLESLLAEYSITDLTAAERDEL
MAB_2863|M.abscessus_ATCC_1997 WRADYFERVSSLTQSADHWTRVQDLYAAGFADVCQTFGLPCPDTATAEDV
Mflv_0739|M.gilvum_PYR-GCK WRAGYVPAMDRVRRGELPWTRIDDLHRGRLVELLDQAAIT-VSDTDIDHL
Mvan_0108|M.vanbaalenii_PYR-1 WRAGYVPAMDRVRRGELPWTRLDDLHRGRLVELLEGTGIT-VSDSEIDEL
MSMEG_3450|M.smegmatis_MC2_155 WRARYRPILDEVNDGLRPWGNFDELHLATLQDVLSERDIA-LSPDQLTSL
*** * :. . * . .:: : : :. :
MMAR_3371|M.marinum_M TLAWQRLRPWPDVVAGLTRLKTKFTLATLSNADVSAVVNISKRAALPWDA
MUL_1268|M.ulcerans_Agy99 TLAWQRLRPWPDVVAGLTRLKTKFTLATLSNADVSAVVNISKRAALPWDA
MAV_2690|M.avium_104 TLAWQRLVPWPDVLPGLTRLKKKFIIATLSNADVSALINIAKRGGLPWDA
MAB_2863|M.abscessus_ATCC_1997 ASSWQRLVPWPDVPAGLAALRMRAVVATLSNTDMATMVNLFKRLDISWDA
Mflv_0739|M.gilvum_PYR-GCK NRAWHRLDPWPDSVAGLTRLKQRFLITTLSNGNVSLLTDMAKRAGLPWDC
Mvan_0108|M.vanbaalenii_PYR-1 NRAWHRLDPWPDAVAGLTRLKRRFVITTLSNGNVSLLTNMAKRAGLPWDC
MSMEG_3450|M.smegmatis_MC2_155 VHAWHRLDPWPDVRDGLELLRSNFLIAPLSNGHVRLVVDLARHGDLRFDC
:*:** **** ** *: . ::.*** .: : :: :: : :*.
MMAR_3371|M.marinum_M IFAAEMAGVFKPDPAAYRMAVGYLGCEPAEVMMVASHKYDLRAAARLGLR
MUL_1268|M.ulcerans_Agy99 IFAAEMAGVFKPDPAAYRMAVGYLGCEPAEVMMVASHKYDLRAAARLGLR
MAV_2690|M.avium_104 VFAAEMAGVFKPDPAIYHMAARYLGLAPRQIMMVASHKYDIRAAASLGFQ
MAB_2863|M.abscessus_ATCC_1997 IFTAEVFGRFKPDPSVYRGALRYLGVEPYEAAMVAAHPYDLRAARQLGMG
Mflv_0739|M.gilvum_PYR-GCK VLSAEIFGHYKPDREAYLGTAHILDVAPGELMMVAAHPSDLRAARDAGLV
Mvan_0108|M.vanbaalenii_PYR-1 VLSAEIFGHYKPDREAYLGCAEILDVAPGELMLVAAHPSDLRGARRAGLA
MSMEG_3450|M.smegmatis_MC2_155 ILSAELAHAYKPAPETYLTAAHLLDVDPSEVMLVAAHPSDLAGARAAGLR
:::**: :** * *. * : :**:* *: .* *:
MMAR_3371|M.marinum_M TAFVARPLEFG--IGGAADATYADEFDINAADFLD---LAGQLGC-----
MUL_1268|M.ulcerans_Agy99 TAFVARPLEFGSVVRPMQPMPMSSTSTPQTSSILPGNSVADRCGVETSFT
MAV_2690|M.avium_104 TAFVTRPFEFG--PLGLADTAYDDAFDVNAADLLD---LATQLSC-----
MAB_2863|M.abscessus_ATCC_1997 TIFVSRPHEYG--DPVLAHTDPDEEFDQRVTAIGE---LS----------
Mflv_0739|M.gilvum_PYR-GCK TAYVHRPLEQG--PDRTPERPGHDEFDITASDFGD---FADQLGA-----
Mvan_0108|M.vanbaalenii_PYR-1 TAFVHRPLEQG--PDRAPARPADGEFDVMADDFHD---LADRLGV-----
MSMEG_3450|M.smegmatis_MC2_155 TAFIDRPLEHG--P-ATPKR-SDRKADVSAADLHE---LARLLGDEGKA-
* :: ** * * . : .:
MMAR_3371|M.marinum_M -----
MUL_1268|M.ulcerans_Agy99 RTEFG
MAV_2690|M.avium_104 -----
MAB_2863|M.abscessus_ATCC_1997 -----
Mflv_0739|M.gilvum_PYR-GCK -----
Mvan_0108|M.vanbaalenii_PYR-1 -----
MSMEG_3450|M.smegmatis_MC2_155 -----