For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPVLEDWHAWAHLFTDDATYFDHFYGTFTGPTEITKFLEGTMGAAPQVYSPLVWYIVDGTRVAYKVLNR ADNPQPGGLPIDFPSYQFIEYAGGGKWRSEEDVWIPAEMKQFAKRYATAADAHPQTLQQKLRREDWGCWV DWARPEPGHKAAPSWLDKAGFTPFSGIADIDFGVRSH*
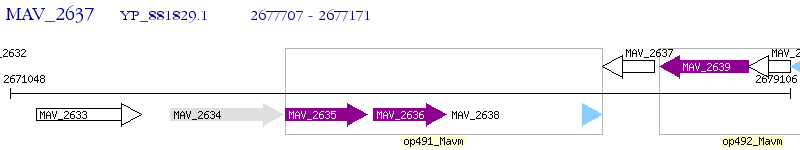
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2637 | - | - | 100% (178) | hypothetical protein MAV_2637 |
| M. avium 104 | MAV_1936 | - | 1e-95 | 85.39% (178) | hypothetical protein MAV_1936 |
| M. avium 104 | MAV_2774 | - | 7e-14 | 35.92% (103) | hypothetical protein MAV_2774 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0143 | - | 7e-10 | 27.50% (120) | hypothetical protein Mb0143 |
| M. gilvum PYR-GCK | Mflv_1107 | - | 9e-13 | 29.75% (121) | hypothetical protein Mflv_1107 |
| M. tuberculosis H37Rv | Rv0138 | - | 1e-10 | 28.33% (120) | hypothetical protein Rv0138 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1928c | - | 1e-11 | 28.93% (121) | hypothetical protein MAB_1928c |
| M. marinum M | MMAR_2850 | - | 5e-14 | 35.92% (103) | hypothetical protein MMAR_2850 |
| M. smegmatis MC2 155 | MSMEG_6475 | - | 2e-10 | 29.75% (121) | hypothetical protein MSMEG_6475 |
| M. thermoresistible (build 8) | TH_0079 | - | 5e-11 | 29.75% (121) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2907 | - | 8e-14 | 35.92% (103) | hypothetical protein MUL_2907 |
| M. vanbaalenii PYR-1 | Mvan_5706 | - | 1e-12 | 29.75% (121) | hypothetical protein Mvan_5706 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6475|M.smegmatis_MC2_155 ---MSRFSRDELSAAFARFEATVDTAARTKDWDVWVRHYTPDVDYVEHAA
TH_0079|M.thermoresistible__bu -VTTPQFSREELAEAFANFEATVDRAARTKDWDPWANQYTVDVTYVEHAA
Mb0143|M.bovis_AF2122/97 -MSASEFSRAELAAAFEKFEKTVARAAATRDWDCWVQHYTPDVEYIEHAA
Rv0138|M.tuberculosis_H37Rv -MSASEFSRAELAAAFEKFEKTVARAAATRDWDCWVQHYTPDVEYIEHAA
Mflv_1107|M.gilvum_PYR-GCK -MTGSTFSRDELVEAFAVFEGTVDRAAQTRDWDPWVDQYTDDILYIEHAA
Mvan_5706|M.vanbaalenii_PYR-1 -MTGNTFTRDELAEAFAGFEKTVDRAAQTRDWDPWVDQYTDDILYIEHAA
MAB_1928c|M.abscessus_ATCC_199 ---MPSFTRAELEEAFAQHQATVHRCIETGDWNPYAEMYTEDALYIEHVV
MMAR_2850|M.marinum_M MSGMGRFTKAEIEQAVRHYTTVVEGCSASGDWRPFADLFTEDVVYTEHHY
MUL_2907|M.ulcerans_Agy99 ---MGRFTKAEIEQAVRHYTTVVEGCSASGDWRPCADLFTEDVVYTEHHY
MAV_2637|M.avium_104 -----------------------MTGPVLEDWHAWAHLFTDDATYFDHFY
** . :* * * :*
MSMEG_6475|M.smegmatis_MC2_155 GTMKGREEVRAWIWKTMTSFPGNHMTAFPSLWTVIDEPTGRVICELDNPM
TH_0079|M.thermoresistible__bu GTMRSREEVREWIWRTMTTFPGSHMTAFPSLWSVIDEPTGRIICELDNPM
Mb0143|M.bovis_AF2122/97 GIMRGRQRVRAWIQETMTTFPGSHMVAFPSLWSVIDESTGRIICELGNPM
Rv0138|M.tuberculosis_H37Rv GIMRGRQRVRAWIQETMTTFPGSHMVAFPSLWSVIDESTGRIICELDNPM
Mflv_1107|M.gilvum_PYR-GCK GTMRGREQVRPWIWKTMESFPGNYMTSFPSLWHVIDEDTGRIICELDNPM
Mvan_5706|M.vanbaalenii_PYR-1 GTMRGRDEVRPWIWKTMESFPGSYMTSFPSLWSVIDEATARIICELDNPM
MAB_1928c|M.abscessus_ATCC_199 GRLHGKEAIRQYITAVMAEFPGNHMPSLPATWTVFDPEKGWVVCEIDNVM
MMAR_2850|M.marinum_M GVFHGREAVREWIVAVMAPFP---HMRFPSDWTAYDEDNDAVVLMIKNSL
MUL_2907|M.ulcerans_Agy99 GVFHGREAVREWIVAVMAPFP---HMRFPSDWTAYDQDNDAVVLMIKNSL
MAV_2637|M.avium_104 GTFTGPTEITKFLEGTMGAAP---QVYSPLVWYIVDG--TRVAYKVLNRA
* : . : :: .* * * * * : : *
MSMEG_6475|M.smegmatis_MC2_155 VDPGD--GTIISATNISILTYAGDGLWSRQEDVYNPLRFASAAMKWCRKA
TH_0079|M.thermoresistible__bu IDPGD--GTVISATNLSILTYAGDGLWCRQEDVYNPLRFASAAMKWCRKA
Mb0143|M.bovis_AF2122/97 LDPGD--GSVISATNISIITYAGNGQWCRQEDIYNPLRFLRAAMKWCRKA
Rv0138|M.tuberculosis_H37Rv LDPGD--GSVISATNISIITYAGNGQWCRQEDIYNPLRFLRAAMKWCRKA
Mflv_1107|M.gilvum_PYR-GCK RDPGD--GTIISATNISILTYAGDGRWRQQEDIYNPLRFVSAAVKWCRKA
Mvan_5706|M.vanbaalenii_PYR-1 RDPGD--GTIISATNISILTYAGDGKWKRQEDIYNPLRFVSATVKWCRKA
MAB_1928c|M.abscessus_ATCC_199 SDPGD--GQHWGEPNLTVLHYAGDGLWSQQEDAYNPANMVKMVRRWCRAA
MMAR_2850|M.marinum_M DHPTDPNGEPFWFPNWTRLVYAGDGLFCSEEDIYNPDRDAPRVIAAWLLA
MUL_2907|M.ulcerans_Agy99 DHPTDPNGEPFWFPNWTRLVYAGDGLFCSEEDIYNPDRDAPRVIAAWLLA
MAV_2637|M.avium_104 DNPQP-GGLPIDFPSYQFIEYAGGGKWRSEEDVWIPAEMKQFAKRYATAA
.* * .. : ***.* : :** : * . . *
MSMEG_6475|M.smegmatis_MC2_155 AELGTLTDEAAEWM--------AKF--GGRR-------------------
TH_0079|M.thermoresistible__bu RELGTLTDEAAAWM--------EQY--GGRA-------------------
Mb0143|M.bovis_AF2122/97 QELGTLDEDAARWM--------RRH--GGP--------------------
Rv0138|M.tuberculosis_H37Rv QELGTLDEDAARWM--------RRH--GGP--------------------
Mflv_1107|M.gilvum_PYR-GCK EELGTLPDEAAEWM--------AKY--GGKR-------------------
Mvan_5706|M.vanbaalenii_PYR-1 EELGTLPDEAAQWM--------AKFSPGGRGAL-----------------
MAB_1928c|M.abscessus_ATCC_199 EAAGNLPDEAREWL--------AKY--GPRQN------------------
MMAR_2850|M.marinum_M G--GQLASDQILQP--------GP--------------------------
MUL_2907|M.ulcerans_Agy99 G--GQLASDQILQP--------GP--------------------------
MAV_2637|M.avium_104 DAHPQTLQQKLRREDWGCWVDWARPEPGHKAAPSWLDKAGFTPFSGIADI
.:
MSMEG_6475|M.smegmatis_MC2_155 -------
TH_0079|M.thermoresistible__bu -------
Mb0143|M.bovis_AF2122/97 -------
Rv0138|M.tuberculosis_H37Rv -------
Mflv_1107|M.gilvum_PYR-GCK -------
Mvan_5706|M.vanbaalenii_PYR-1 -------
MAB_1928c|M.abscessus_ATCC_199 -------
MMAR_2850|M.marinum_M -------
MUL_2907|M.ulcerans_Agy99 -------
MAV_2637|M.avium_104 DFGVRSH