For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LLRHRRKILCAYSFGERAMGSVAQRQLHRWVVGASGRARSRRCRPSARPADRRPAQAGGAATTADAFLGC VRDGRRLRLVDAVMTQNDVIVARASALFLRRSEHAVDTVWTSPVTMPAVPAEPDVLNDDVPMVFHSFGRD PVAGSPGVGIKEWRHHGQKFAWMRETKFLVDDEPLSPFTRAVMAGDVTSSLTHWGTEGLQFINADYTITL SRLPEGVYIGLASVTHYSHAGVATGVATLFDETGPIGSGMATALSNPGFTPPPSMVVS*
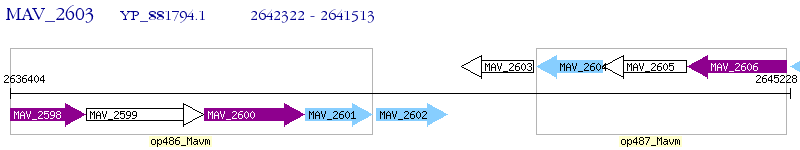
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2603 | - | - | 100% (269) | hypothetical protein MAV_2603 |
| M. avium 104 | MAV_1885 | - | 4e-96 | 83.16% (196) | hypothetical protein MAV_1885 |
| M. avium 104 | MAV_3659 | - | 9e-55 | 54.01% (187) | hypothetical protein MAV_3659 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0790 | - | 2e-53 | 54.69% (192) | hypothetical protein Mflv_0790 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2154c | - | 9e-07 | 28.80% (191) | hypothetical protein MAB_2154c |
| M. marinum M | MMAR_0390 | - | 1e-08 | 30.11% (93) | hypothetical protein MMAR_0390 |
| M. smegmatis MC2 155 | MSMEG_3458 | - | 2e-51 | 51.56% (192) | hypothetical protein MSMEG_3458 |
| M. thermoresistible (build 8) | TH_4503 | - | 3e-50 | 51.04% (192) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2342 | - | 2e-07 | 26.94% (193) | hypothetical protein MUL_2342 |
| M. vanbaalenii PYR-1 | Mvan_0060 | - | 1e-50 | 52.36% (191) | hypothetical protein Mvan_0060 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0790|M.gilvum_PYR-GCK -----MSS----RAFFLPDGDAFAPTDWARGPWGQTISG-NYVGGLLGHV
Mvan_0060|M.vanbaalenii_PYR-1 -----MTT----RAYFLPDRDGFVPTALARGPWGGTISG-NYVGGLLGHV
TH_4503|M.thermoresistible__bu -----MSTSATRAAYFGIDGDRYIPSPLTKGPWGDQLSG-TFIGGLLAHA
MSMEG_3458|M.smegmatis_MC2_155 -----MDRNG-LTPYFMRRDDGFVPNEIAHGGWGPTLGG-HVVGGLLARA
MAV_2603|M.avium_104 -----MLLR-----HRRKILCAYSFGERAMGSVAQRQLH-RWVVGASGRA
MAB_2154c|M.abscessus_ATCC_199 MTLDLTGCYYRRLD-ADGEFQVFESTELTASGWGPNLQHGSPPMALLLKA
MUL_2342|M.ulcerans_Agy99 ----MNGCYYRRLPGADGDYQMFDSTDFTRSNWDPDIQHGSPPLALLTKL
MMAR_0390|M.marinum_M ----MTDSYYELIDDADALGEKFAATDLVRGTWSASIQHGAPVSALLVRA
: . . . :
Mflv_0790|M.gilvum_PYR-GCK IERDTGH-PDLQPARLTVDLCRPATLTEPVSVATTVVREGRRLTLVDAVM
Mvan_0060|M.vanbaalenii_PYR-1 IDRDTGD-PELQPARLTVDLMRPVAM-APVQVQTSVVRSGRRLKLVEASM
TH_4503|M.thermoresistible__bu VERDAWD-ADFHPTRFTVDLLRPVAL-APMRLRTTVVRSSSRLRLVDSEI
MSMEG_3458|M.smegmatis_MC2_155 IERHVDD-AELQPARLTVEILRRVAS-APVRVSASVVRSGSRMKALDAEM
MAV_2603|M.avium_104 RSRRCR--PSARPADRRPAQAGGAATTADAFLG--CVRDGRRLRLVDAVM
MAB_2154c|M.abscessus_ATCC_199 IEELPEPRENLRIGRVALDILGPVPLEP-VRVRAWIERPGRRIALAVAEM
MUL_2342|M.ulcerans_Agy99 IEELSAG-SGLRIGRFSLDILGAIPVAP-VRARAWVQRPGSRVAMMVAEM
MMAR_0390|M.marinum_M LQRCAWR-EDTRLSRVVVDLIGAVPADGDLWVSSRIERGGKQIELVSAEL
.. : . * . :: : :
Mflv_0790|M.gilvum_PYR-GCK TQGG-----NFVARASTLFLRRGEQPPGPRFTTTVAMPEVP-RADSVPDD
Mvan_0060|M.vanbaalenii_PYR-1 TQSG-----TVVARASALMLRRGEQPPGPRWSTPVRMPPVPPLPDPPPKR
TH_4503|M.thermoresistible__bu VQDD-----TVVSRASTLLLRPGDHPPGKVWSTPVDVPPRPSRFPDTGFA
MSMEG_3458|M.smegmatis_MC2_155 TQQG-----EVVARASALYLRRGVQPGGRFWSTPVDLPPIPREPERFDDT
MAV_2603|M.avium_104 TQND-----VIVARASALFLRRSEHAVDTVWTSPVTMPAVPAEPDVLNDD
MAB_2154c|M.abscessus_ATCC_199 EARA-NGGYRAVARASAWLLATSVT------ADKASDR-FPPLPEYPAGP
MUL_2342|M.ulcerans_Agy99 VA------DRVLARVTAWLLAVSDT------TDVASDR-YPPLVEGPAQP
MMAR_0390|M.marinum_M AAPGPDGRPRAVARASGWRLAQLDT------QELRHAAEQPPRPLAEARN
::*.: * *
Mflv_0790|M.gilvum_PYR-GCK IATLVWSYGKDGDTPRMG--LDAWTYAGPKYIWALELAPIVAGTELTPFE
Mvan_0060|M.vanbaalenii_PYR-1 AVLMVWSYGNNGDTPAPG--LDAWRYDGPKRIWTCDLGPLVAGTEQSAFT
TH_4503|M.thermoresistible__bu GGMAFWLYGADLDNPTAGPDFTPWQYVGPKHGWVRELNPLIDGVDLTPFT
MSMEG_3458|M.smegmatis_MC2_155 VPMFITPYGAPPGSIDSP---FPWQYDGPRYAWLREVRDLVDGEELTPLL
MAV_2603|M.avium_104 VPMVFHSFGRDPVAGSPGVGIKEWRHHGQKFAWMRETKFLVDDEPLSPFT
MAB_2154c|M.abscessus_ATCC_199 TPQAFGFEGSNYSKSVDFQWFTVPNGE-PQVAWMRPHVHIVDTEPTTPWQ
MUL_2342|M.ulcerans_Agy99 RPAAFAGVGG-YFDALSWRPQDTEGQP-AAVSWFRPTAHIVDTDPASPLQ
MMAR_0390|M.marinum_M RKLAPMSWDRNYVHSLDWRWLTEPMTPGPGECWIKPTVDLVNGETMTPLE
. * :: :.
Mflv_0790|M.gilvum_PYR-GCK RAALAGDMASSL-THFGEDGLPFINADYTLTLSRLPDGPYLGLAALTHDS
Mvan_0060|M.vanbaalenii_PYR-1 RAAVVGEMASSL-THFGVDGLPFINADYTLTLSRLPETPYLGLAALTHFS
TH_4503|M.thermoresistible__bu RAALAGDITSSL-TQFGDAGLYYINPDYTLTLSRLPDGPDIGLSAVTHYS
MSMEG_3458|M.smegmatis_MC2_155 RAALAVDVTSSL-TNFSTDGLAFINADYTLTLSRLPRGLYIGMAALVHAS
MAV_2603|M.avium_104 RAVMAGDVTSSL-THWGTEGLQFINADYTITLSRLPEGVYIGLASVTHYS
MAB_2154c|M.abscessus_ATCC_199 RLASVIDSANGIGAVLNPLRFVYMNTDTVMHVHRIPQGDEFGVRARMSVG
MUL_2342|M.ulcerans_Agy99 QLAAVVDCANGVGAILDPNRFLFMNTDTVVHLHRLPTGTDFALRARASVG
MMAR_0390|M.marinum_M RLFAVADCANGIGSKLDITKWTFLNTDLAVHVFRIPAGDWIGIRAETSYG
: . : :..: : . ::*.* .: : *:* :.: : .
Mflv_0790|M.gilvum_PYR-GCK AAGVATGTAIVVDETGPLGTATATALANPGFSPP-----------RAFS-
Mvan_0060|M.vanbaalenii_PYR-1 HAGVATGTAVVVDELGPIGTATATAVANPGFDPA-----------HPIG-
TH_4503|M.thermoresistible__bu HAGIATGVATLFDRRGPIGSAMATGLAHGGFRAPTLDEVEKLGGIRPNGA
MSMEG_3458|M.smegmatis_MC2_155 AEGVATGTAALFDQDGPIGTGVSTAVANFNFTPN-------VEVDKVTGE
MAV_2603|M.avium_104 HAGVATGVATLFDETGPIGSGMATALSNPGFTPP-----------PSMVV
MAB_2154c|M.abscessus_ATCC_199 PDGVGVTTAEIFDRQGHIGSSAQTVLVQRR--------------------
MUL_2342|M.ulcerans_Agy99 PDGLGVTTAEIFDTTGFVGTSAQTLLVRRR--------------------
MMAR_0390|M.marinum_M PDGIGTAAGTLFDQQGAVGTIAQSVLVRRRAR------------------
*:.. .. :.* * :*: : : .
Mflv_0790|M.gilvum_PYR-GCK ---
Mvan_0060|M.vanbaalenii_PYR-1 ---
TH_4503|M.thermoresistible__bu AG-
MSMEG_3458|M.smegmatis_MC2_155 SRI
MAV_2603|M.avium_104 S--
MAB_2154c|M.abscessus_ATCC_199 ---
MUL_2342|M.ulcerans_Agy99 ---
MMAR_0390|M.marinum_M ---