For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGRAVEQQVDDVDLQPARLTVDLLRPVALQPVQVTSSVVRHGRRIRLVDAVMRQNDAIVARASALYLRRS EHRADQVWTSPVSMPAIPAEPVVVPDDLPMMLHSYGRDPVAGSPGVGVTEWRHDGQKFAWVRETKLLVDD EPLSPFTRAVMAGDVTSSLTHWGTAGLQFINADYTVSLSRLPQGVYIGLASVTHYDHGGVATGVATLFDE AGPIGSGLATALANPGFTPPSSMLST*
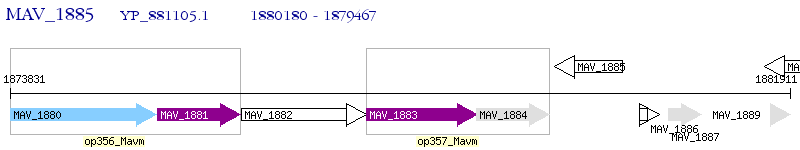
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1885 | - | - | 100% (237) | hypothetical protein MAV_1885 |
| M. avium 104 | MAV_2603 | - | 3e-96 | 83.16% (196) | hypothetical protein MAV_2603 |
| M. avium 104 | MAV_3659 | - | 6e-68 | 55.95% (227) | hypothetical protein MAV_3659 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0790 | - | 2e-62 | 52.59% (232) | hypothetical protein Mflv_0790 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2154c | - | 1e-13 | 30.17% (232) | hypothetical protein MAB_2154c |
| M. marinum M | MMAR_0390 | - | 6e-14 | 27.35% (234) | hypothetical protein MMAR_0390 |
| M. smegmatis MC2 155 | MSMEG_3458 | - | 2e-65 | 54.11% (231) | hypothetical protein MSMEG_3458 |
| M. thermoresistible (build 8) | TH_4503 | - | 5e-61 | 50.86% (232) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2342 | - | 1e-10 | 25.75% (233) | hypothetical protein MUL_2342 |
| M. vanbaalenii PYR-1 | Mvan_0060 | - | 5e-64 | 52.61% (230) | hypothetical protein Mvan_0060 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0790|M.gilvum_PYR-GCK -----MSS----RAFFLPDGDAFAPTDWARGPWGQTIS-GNYVGGLLGHV
Mvan_0060|M.vanbaalenii_PYR-1 -----MTT----RAYFLPDRDGFVPTALARGPWGGTIS-GNYVGGLLGHV
TH_4503|M.thermoresistible__bu -----MSTSATRAAYFGIDGDRYIPSPLTKGPWGDQLS-GTFIGGLLAHA
MAV_1885|M.avium_104 ---------------------------------------------MLGRA
MSMEG_3458|M.smegmatis_MC2_155 ------MDRNGLTPYFMRRDDGFVPNEIAHGGWGPTLG-GHVVGGLLARA
MAB_2154c|M.abscessus_ATCC_199 MTLDLTGCYYRRLD-ADGEFQVFESTELTASGWGPNLQHGSPPMALLLKA
MUL_2342|M.ulcerans_Agy99 ----MNGCYYRRLPGADGDYQMFDSTDFTRSNWDPDIQHGSPPLALLTKL
MMAR_0390|M.marinum_M ----MTDSYYELIDDADALGEKFAATDLVRGTWSASIQHGAPVSALLVRA
:* :
Mflv_0790|M.gilvum_PYR-GCK IERDTGH-PDLQPARLTVDLCRPATLTEPVSVATTVVREGRRLTLVDAVM
Mvan_0060|M.vanbaalenii_PYR-1 IDRDTGD-PELQPARLTVDLMRPVAM-APVQVQTSVVRSGRRLKLVEASM
TH_4503|M.thermoresistible__bu VERDAWD-ADFHPTRFTVDLLRPVAL-APMRLRTTVVRSSSRLRLVDSEI
MAV_1885|M.avium_104 VEQQVDD-VDLQPARLTVDLLRPVAL-QPVQVTSSVVRHGRRIRLVDAVM
MSMEG_3458|M.smegmatis_MC2_155 IERHVDD-AELQPARLTVEILRRVAS-APVRVSASVVRSGSRMKALDAEM
MAB_2154c|M.abscessus_ATCC_199 IEELPEPRENLRIGRVALDILGPVPLEP-VRVRAWIERPGRRIALAVAEM
MUL_2342|M.ulcerans_Agy99 IEELSAG-SGLRIGRFSLDILGAIPVAP-VRARAWVQRPGSRVAMMVAEM
MMAR_0390|M.marinum_M LQRCAWR-EDTRLSRVVVDLIGAVPADGDLWVSSRIERGGKQIELVSAEL
::. : *. ::: . : : : * . :: : :
Mflv_0790|M.gilvum_PYR-GCK TQGG-----NFVARASTLFLRRGEQPPGPRFTTTVAMPEVP-RADSVPDD
Mvan_0060|M.vanbaalenii_PYR-1 TQSG-----TVVARASALMLRRGEQPPGPRWSTPVRMPPVPPLPDPPPKR
TH_4503|M.thermoresistible__bu VQDD-----TVVSRASTLLLRPGDHPPGKVWSTPVDVPPRPSRFPDTGFA
MAV_1885|M.avium_104 RQND-----AIVARASALYLRRSEHRADQVWTSPVSMPAIPAEPVVVPDD
MSMEG_3458|M.smegmatis_MC2_155 TQQG-----EVVARASALYLRRGVQPGGRFWSTPVDLPPIPREPERFDDT
MAB_2154c|M.abscessus_ATCC_199 EARA-NGGYRAVARASAWLLATSVTADKASDR-FPPLPEYPAGPTPQAFG
MUL_2342|M.ulcerans_Agy99 VA------DRVLARVTAWLLAVSDTTDVASDR-YPPLVEGPAQPRPAAFA
MMAR_0390|M.marinum_M AAPGPDGRPRAVARASGWRLAQLDTQELRHAAEQPPRPLAEARNRKLAPM
::*.: *
Mflv_0790|M.gilvum_PYR-GCK IATLVWSYGKDG-DTPRMG--LDAWTYAGPKYIWALELAPIVAGTELTPF
Mvan_0060|M.vanbaalenii_PYR-1 AVLMVWSYGNNG-DTPAPG--LDAWRYDGPKRIWTCDLGPLVAGTEQSAF
TH_4503|M.thermoresistible__bu GGMAFWLYGADL-DNPTAGPDFTPWQYVGPKHGWVRELNPLIDGVDLTPF
MAV_1885|M.avium_104 LPMMLHSYGRDP-VAGSPGVGVTEWRHDGQKFAWVRETKLLVDDEPLSPF
MSMEG_3458|M.smegmatis_MC2_155 VPMFITPYGAPPGSIDSP----FPWQYDGPRYAWLREVRDLVDGEELTPL
MAB_2154c|M.abscessus_ATCC_199 FEGSNYSKSVDFQWFTVP-------NGE-PQVAWMRPHVHIVDTEPTTPW
MUL_2342|M.ulcerans_Agy99 GVGG-YFDALSWRPQDTE-------GQP-AAVSWFRPTAHIVDTDPASPL
MMAR_0390|M.marinum_M SWDRNYVHSLDWRWLTEP-------MTPGPGECWIKPTVDLVNGETMTPL
. * :: :.
Mflv_0790|M.gilvum_PYR-GCK ERAALAGDMASSL-THFGEDGLPFINADYTLTLSRLPDGPYLGLAALTHD
Mvan_0060|M.vanbaalenii_PYR-1 TRAAVVGEMASSL-THFGVDGLPFINADYTLTLSRLPETPYLGLAALTHF
TH_4503|M.thermoresistible__bu TRAALAGDITSSL-TQFGDAGLYYINPDYTLTLSRLPDGPDIGLSAVTHY
MAV_1885|M.avium_104 TRAVMAGDVTSSL-THWGTAGLQFINADYTVSLSRLPQGVYIGLASVTHY
MSMEG_3458|M.smegmatis_MC2_155 LRAALAVDVTSSL-TNFSTDGLAFINADYTLTLSRLPRGLYIGMAALVHA
MAB_2154c|M.abscessus_ATCC_199 QRLASVIDSANGIGAVLNPLRFVYMNTDTVMHVHRIPQGDEFGVRARMSV
MUL_2342|M.ulcerans_Agy99 QQLAAVVDCANGVGAILDPNRFLFMNTDTVVHLHRLPTGTDFALRARASV
MMAR_0390|M.marinum_M ERLFAVADCANGIGSKLDITKWTFLNTDLAVHVFRIPAGDWIGIRAETSY
: . : :..: : . ::*.* .: : *:* :.: :
Mflv_0790|M.gilvum_PYR-GCK SAAGVATGTAIVVDETGPLGTATATALANPGFSPP-----------RAFS
Mvan_0060|M.vanbaalenii_PYR-1 SHAGVATGTAVVVDELGPIGTATATAVANPGFDPA-----------HPIG
TH_4503|M.thermoresistible__bu SHAGIATGVATLFDRRGPIGSAMATGLAHGGFRAPTLDEVEKLGGIRPNG
MAV_1885|M.avium_104 DHGGVATGVATLFDEAGPIGSGLATALANPGFTPPS----------SMLS
MSMEG_3458|M.smegmatis_MC2_155 SAEGVATGTAALFDQDGPIGTGVSTAVANFNFTPNV----------EVDK
MAB_2154c|M.abscessus_ATCC_199 GPDGVGVTTAEIFDRQGHIGSSAQTVLVQRR-------------------
MUL_2342|M.ulcerans_Agy99 GPDGLGVTTAEIFDTTGFVGTSAQTLLVRRR-------------------
MMAR_0390|M.marinum_M GPDGIGTAAGTLFDQQGAVGTIAQSVLVRRRAR-----------------
. *:.. .. :.* * :*: : :..
Mflv_0790|M.gilvum_PYR-GCK -------
Mvan_0060|M.vanbaalenii_PYR-1 -------
TH_4503|M.thermoresistible__bu AAG----
MAV_1885|M.avium_104 T------
MSMEG_3458|M.smegmatis_MC2_155 VTGESRI
MAB_2154c|M.abscessus_ATCC_199 -------
MUL_2342|M.ulcerans_Agy99 -------
MMAR_0390|M.marinum_M -------