For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTTQKPSADAGASEATGDDLAAEEGRITDEDIERAKGQIGIPVHQRDEAWNKLPSADAITHFAFGCGDD NPLFHDPTYGLSTRWHGQIASPTFPIATGLDQTPKFTDPERKKLFRGLFRGTGKYYSGVKWTWYRPIYAG RPVLAENYTLDVQVKESEFSGGRSVKETFRYLYVDIDGNPIATRDESYINAERQGSKKSGKLKDIARKHW TPEEFAQVEEEYEAERRRGADPQWWEDVSVGDELPGIIKGPLTVVDIISMHMGWGWGGYGVGPLKFAHQL RKRMPAFYQPDEYGVPDVVQRLHWDAERAQALGIPAPYDYGQMRAAWVSHLLTNWIGDDGWLAEMDLQLR GFNYHGDVHRCTGTVTSKGEGAEDPVSLDVFATSQRGETTTRGTAKVLLPSKATGAVVLPIPDADLRRRG AQVVSRVSGKVGEEMRRLYGE
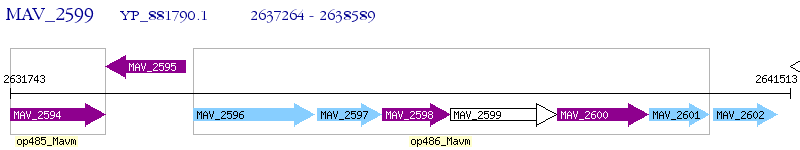
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2599 | - | - | 100% (441) | hypothetical protein MAV_2599 |
| M. avium 104 | MAV_1882 | - | 0.0 | 82.54% (441) | hypothetical protein MAV_1882 |
| M. avium 104 | MAV_1946 | - | 2e-09 | 32.04% (103) | MaoC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2055 | - | 3e-05 | 28.87% (97) | MaoC-like dehydratase |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3972 | - | 2e-89 | 43.62% (392) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2599|M.avium_104 MTTTQKPSADAGASEATGDDLAAEEGRITDEDIERAKGQIGIPVHQRDEA
TH_3972|M.thermoresistible__bu ---------------------VTATGRITDEGLARLRATIGIAVPHPQPP
MAB_2055|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2599|M.avium_104 WNKLPSADAITHFAFGCGDDNPLFHDPTYGLSTRWHGQIASPTFP---IA
TH_3972|M.thermoresistible__bu HYRRPNEDCFRQVAECYGDPNPLWCDPEYAAKTRWDGSIAPPALVGGDTL
MAB_2055|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2599|M.avium_104 TGLDQTPKFTDPERKKLFRGLFRGTGKYYSGVKWTWYRPIYAGRPVLAEN
TH_3972|M.thermoresistible__bu VGENEVTTLSDRERELTKGDPLRGVHAFYASSMREWWAPLYADRRVFRRN
MAB_2055|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2599|M.avium_104 YTLDVQVKESEFSGGRSVKETFRYLYVDIDGNPIATRDESYINAERQGSK
TH_3972|M.thermoresistible__bu ALVAALDKRSDFAG-RAVHEWTAEVFRDEAGTLLGAQYRNMIRTERSRAR
MAB_2055|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2599|M.avium_104 KSGKLKDIARKHWTPEEFAQVEEEYEAERRRGADPQWWEDVSVGDELPGI
TH_3972|M.thermoresistible__bu SRRKYDDIEPKTWTPEEIAEIDARYAAEHPRGAEPRWWEDVAEGDTLGPL
MAB_2055|M.abscessus_ATCC_1997 ----------------------------MTTSPAP---VALAAGDEMPSS
.. * :: ** :
MAV_2599|M.avium_104 IKGPLTVVDIISMHMGWGWGGYGVGPLKFAHQLRKRMPAFYQPDEYGVPD
TH_3972|M.thermoresistible__bu TKGPLTVTDMVCWHVGMGMGLYGVRPLRLAWQNRRRIPRFYTPDAQGVPD
MAB_2055|M.abscessus_ATCC_1997 QFGPLTRSHIVRYAGSGGD---------------------FNP-------
**** .:: . * : *
MAV_2599|M.avium_104 VVQRLHWDAERAQALGIPAPYDYGQMRAAWVSHLLTNWIGDDGWLAEMDL
TH_3972|M.thermoresistible__bu VAQRVHWDPGAARDVGNPTTFDYGRMRETWLIHLCTDWMGDDGWLWRLDC
MAB_2055|M.abscessus_ATCC_1997 ----IHHDEEFARAAGMPGVFGMGLLHGGVLAQRLTAWVG-LANIRSFRI
:* * *: * * :. * :: : : * *:* . : :
MAV_2599|M.avium_104 QLRGFNYHGDVHRCTGTVTSK--GEGAEDPVSLDVFATSQRGETTTRGTA
TH_3972|M.thermoresistible__bu EFRRFNYVGDLHTITGTVARKYLADGDRPAVDLDLAATNQRGEVTTPGHA
MAB_2055|M.abscessus_ATCC_1997 RFTGQVWPGDLLSFGGRVTGVR-EAGGQQVADLELVVTRQSGEPAIKGSA
.: : **: * *: * . ..*:: .* * ** : * *
MAV_2599|M.avium_104 KVLLPSKATGAVVLPIPDADLRRRGAQVVSRVSGKVGEEMRRLYGE
TH_3972|M.thermoresistible__bu TVLLPSRRHGPVRLPDPPG-----GADTLTGVLQAVIAEFEHR---
MAB_2055|M.abscessus_ATCC_1997 VVQVAR----------------------------------------
* :.