For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHSHRLVDHIVGYLASIGVDCIFGVDGANIEDLYDAAFLHSDMDAVLAKHEFSAVAMADGYSRSGSGFG VVATSAWGALNLVAGLGELFTSRVPLLALVGQPPSTMDGRGGFQDTSGRNGSPNAEALFSEVSVLCRRVQ TPADIALALPAAVAAARTGGPAVLLLPKDIQQADVTSNGYLPEPGIERPRDIGDPRPIVAALRRASGPVT IIAGEQVVRDDARAELASLRAVLRARVACVPHAKDAAGTPGFGLSSALGVTGIMGHPGVVEAVADSALCL VVGTRLAVTARAGLDDALAAVPTISLGSAPPYLACTHIHTDDLRASLRMLTSALTGRERPTGIRVPDAVS RTELNPPRFDGPGVRYREAMSMLDRVLPDGSDIVVDVGSCGAAAVHYLPVRRGGRFVVAPGMGYSFGAGI GMAVRRRKFGRRGHVVVIAGDGAFFMHGMEVHTAVQYRLPVAFVLINNNAQAMCAMSEKLCYGNRHSYNT FAPSRLGAGLAAMFPELPSVDVDDVDGFGAAVQSALEVDGPSVIGVECSAEEVPPVSAFLGPVTPKSVEC QHGMAQDRIDAIVGA
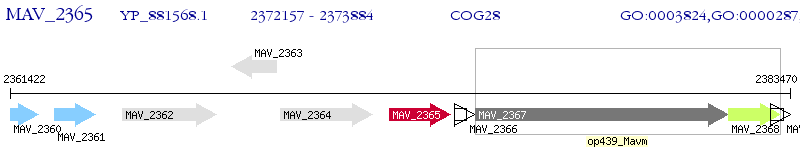
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2365 | - | - | 100% (575) | acetolactate synthase |
| M. avium 104 | MAV_4424 | - | 0.0 | 65.35% (583) | acetolactate synthase |
| M. avium 104 | MAV_4093 | - | 5e-19 | 27.27% (583) | pyruvate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3499c | ilvB2 | 2e-27 | 26.06% (568) | acetolactate synthase large subunit |
| M. gilvum PYR-GCK | Mflv_3445 | - | 1e-22 | 26.83% (589) | pyruvate decarboxylase |
| M. tuberculosis H37Rv | Rv3470c | ilvB2 | 2e-28 | 26.23% (568) | acetolactate synthase large subunit |
| M. leprae Br4923 | MLBr_01696 | ilvB | 1e-22 | 25.05% (511) | acetolactate synthase 1 catalytic subunit |
| M. abscessus ATCC 19977 | MAB_2165 | - | 1e-133 | 47.90% (572) | acetolactate synthase large subunit |
| M. marinum M | MMAR_1710 | ilvB1 | 3e-23 | 24.90% (506) | acetolactate synthase (large subunit) IlvB1 |
| M. smegmatis MC2 155 | MSMEG_1488 | - | 0.0 | 64.70% (558) | acetolactate synthase |
| M. thermoresistible (build 8) | TH_3455 | - | 0.0 | 63.39% (579) | PUTATIVE thiamine pyrophosphate enzyme domain protein |
| M. ulcerans Agy99 | MUL_1947 | ilvB1 | 5e-23 | 24.70% (506) | acetolactate synthase 1 catalytic subunit |
| M. vanbaalenii PYR-1 | Mvan_1352 | - | 1e-174 | 60.73% (550) | thiamine pyrophosphate binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1488|M.smegmatis_MC2_155 ----------------------------------------------MVDH
Mvan_1352|M.vanbaalenii_PYR-1 -----------------------------MAARSTDG-------YRVVDH
TH_3455|M.thermoresistible__bu -----------------------------MAFNGADTPGGPLGRMRVVDY
MAV_2365|M.avium_104 -----------------------------MSHS-----------HRLVDH
MAB_2165|M.abscessus_ATCC_1997 ----------------------------------------------MVDW
Mb3499c|M.bovis_AF2122/97 --------------------------------------------MTVGDH
Rv3470c|M.tuberculosis_H37Rv --------------------------------------------MTVGDH
MMAR_1710|M.marinum_M MSAPTKRHSP-----TSEPQPRGVAAGVNS--SPVHSKHVAPEQLTGAQS
MUL_1947|M.ulcerans_Agy99 MSAPTKRHSP-----TSEPQPRGVAAGVNS--SLVHSKHVAPEQLTGAQS
MLBr_01696|M.leprae_Br4923 MSAPTKPHARPQGAGNSVPNTVKPATQFPSKPAVAKLERVSPEQLTGAQS
Mflv_3445|M.gilvum_PYR-GCK ----------------------------------------MTMSTDVADF
:
MSMEG_1488|M.smegmatis_MC2_155 IVGCLAQSGISHIFGVDGANIEDLYDAAFFRDD-ITAVLAKHEFSAAAMA
Mvan_1352|M.vanbaalenii_PYR-1 IVGTLAAHGVRHIFGVDGANIEDLYDAAYFDDV-VTAVLAKHEFSAAAMA
TH_3455|M.thermoresistible__bu ILDYLAAGAVTHIFGVDGANIEDVYDAAYFHDG-ITAVLAKHEFSAATMA
MAV_2365|M.avium_104 IVGYLASIGVDCIFGVDGANIEDLYDAAFLHSD-MDAVLAKHEFSAVAMA
MAB_2165|M.abscessus_ATCC_1997 LVSELRRHAVDFVFGVDGANIEDLYDAVHYAPG-MQAVVAKHEFSAGTMA
Mb3499c|M.bovis_AF2122/97 LVARMRAAGISVVCGLPTSRLDSLLVRLSRDAG-FQIVLARHEGGAGYLA
Rv3470c|M.tuberculosis_H37Rv LVARMRAAGISVVCGLPTSRLDSLLVRLSRDAG-FQIVLARHEGGAGYLA
MMAR_1710|M.marinum_M VIRSLEELDVEIIFGIPGGAVLPVYDPLFDSQK-LRHVLVRHEQGAGHAA
MUL_1947|M.ulcerans_Agy99 VIRSLEELDVEIIFGIPGGAVLPVYDPLFDSQK-LRHVLVRHEQGAGHAA
MLBr_01696|M.leprae_Br4923 VIRSLEELDVEVIFGIPGGAVLLVYDPLFYSKK-LRHVLVRHEQGAGHAA
Mflv_3445|M.gilvum_PYR-GCK ILDRLRQWNVTHIFGYPGDGINGLVSALGRADDDPMFVQTRHEEMAAFAA
:: : : : * : : * .:** * *
MSMEG_1488|M.smegmatis_MC2_155 DGYSRSGAGIGVVAATSGGGCLNTVPGLGESLASRVPVLALIGQPPTTLD
Mvan_1352|M.vanbaalenii_PYR-1 DGYSRSGAGLGVVAATSGGGCLNTVAALGESLASRVPVLALIGQAPTGLD
TH_3455|M.thermoresistible__bu DGYSRSGAGLGVVAATSGGGSLNLVAALGESFASRVPVLALVGQPPTTLD
MAV_2365|M.avium_104 DGYSRSGSGFGVVATSAWG-ALNLVAGLGELFTSRVPLLALVGQPPSTMD
MAB_2165|M.abscessus_ATCC_1997 DGYARVTSRMSVVATTSGGGAVNLVPALAESYASAVPVLAIVGQPPRSLE
Mb3499c|M.bovis_AF2122/97 DGFARASGKS-AAVFVAGPGATNVISAVANASVNQVPMLILTGEVAVGEF
Rv3470c|M.tuberculosis_H37Rv DGFARASGKS-AAVFVAGPGATNVISAVANASVNQVPMLILTGEVAVGEF
MMAR_1710|M.marinum_M SGYAHATGKVGVCMATSGPGATNLVTPLADAQMDSIPVVAITGQVGRGLI
MUL_1947|M.ulcerans_Agy99 SGYAHATGKVGVCMATSGPGATNLVTPLADAQMDSIPVVAITGQVGRGLI
MLBr_01696|M.leprae_Br4923 SGYAHVTGKVGVCMATSGPGATNLVTPLADAQMDSVPVVAITGQVGRSLI
Mflv_3445|M.gilvum_PYR-GCK VGYAKFSGEVGVCLATSGPGAIHLLNGLYDAKLDHVPVVAIIGQTARTAM
*::: . . : . : : : : . :*:: : *:
MSMEG_1488|M.smegmatis_MC2_155 GQGSFQDTSGDNGALDALSLFSAVSVYCRRVLSPDAIHTVLPEALAAART
Mvan_1352|M.vanbaalenii_PYR-1 GRGAFQDTSGVNGALDALALFSAVSVHCRRITRAEDVRTALPQALAAAAG
TH_3455|M.thermoresistible__bu GRGSFQDTSGRNGALDAEALFTTVSVLCERVCSAADIVAELPRAIMAALT
MAV_2365|M.avium_104 GRGGFQDTSGRNGSPNAEALFSEVSVLCRRVQTPADIALALPAAVAAART
MAB_2165|M.abscessus_ATCC_1997 GNGAFQDGSGRAGSMDLRAVFRPVSGYCARVEHSGEIAEALSGALAAIRD
Mb3499c|M.bovis_AF2122/97 GLHSQQDTS--DDGLGLGATFRRFCRCSVSIESIANARSKIDSAFRALAS
Rv3470c|M.tuberculosis_H37Rv GLHSQQDTS--DDGLGLGATFRRFCRCSVSIESIANARSKIDSAFRALAS
MMAR_1710|M.marinum_M GTDAFQEAD--ISGITMPITKHNFLVRTG-----DDIPRVLAEAFHIAAS
MUL_1947|M.ulcerans_Agy99 GTDAFQEAD--ISGITMPITKHNFLVRTG-----DDIPRVLAEAFHIAAS
MLBr_01696|M.leprae_Br4923 GTDAFQEAD--ISGITMPITKHNFLVRAG-----DDIPRVLAEAFHIASS
Mflv_3445|M.gilvum_PYR-GCK GGSYQQEVD-------LQALFGDVAVYVVEINASAQLPFALDRAIRTARS
* *: . . : *.
MSMEG_1488|M.smegmatis_MC2_155 G--GPAVLLLPKDVQQAVLGEHVNGHVPVMQLG--------TRISELTMM
Mvan_1352|M.vanbaalenii_PYR-1 G--GPAVLLLPKDVQQAALGR--NGVEIVTASR--------TAHSDLSAI
TH_3455|M.thermoresistible__bu G--GPAVLLLPKDVQQAQLNRQHHDDRQAEHAAGR-PR---VRFSDPHPI
MAV_2365|M.avium_104 G--GPAVLLLPKDIQQADVTSNGYLPEPGIERP--------RDIGDPRPI
MAB_2165|M.abscessus_ATCC_1997 G--LPAVLLIPKDVQQARLAVEPQLREPLPRLG--------TVPVQLADF
Mb3499c|M.bovis_AF2122/97 IPRGPVHIALPRDLVDERLPAHQLGTAAAGLGGLRTLA---PCGPDVADE
Rv3470c|M.tuberculosis_H37Rv IPRGPVHIALPRDLVDERLPAHQLGTAAAGLGGLRTLA---PCGPDVADE
MMAR_1710|M.marinum_M GRPGAVLVDIPKDVLQAQCTFSWP--PQMDLPGYKPNT---KPHNRQIRE
MUL_1947|M.ulcerans_Agy99 GRPGAVLVDIPKDVLQAQCTFSWP--PQMDLPGYKPNT---KPHNRQIRE
MLBr_01696|M.leprae_Br4923 GRPGAVLVDIPKDVLQGQCKFSWP--PKMDLPGYKPNT---KPHNRQIRA
Mflv_3445|M.gilvum_PYR-GCK RR-APTVVIVPSDLQEEEYTPPEHQFKHVPSSDPGDVVGVVTPSDAQVRR
.. : :* *: :
MSMEG_1488|M.smegmatis_MC2_155 VRALRRADGPITIFAGEQVARDDARAELERLRATLRARIATVPDAKDVGG
Mvan_1352|M.vanbaalenii_PYR-1 ISALRRVDGPVTIIAGEQVARDDARPHLEQLRDALGAQVATVPDAKDVAR
TH_3455|M.thermoresistible__bu ARALQQARGPVTIIAGEQVARDDARRELEMLRATLRARVACVPDAKDVAG
MAV_2365|M.avium_104 VAALRRASGPVTIIAGEQVVRDDARAELASLRAVLRARVACVPHAKDAAG
MAB_2165|M.abscessus_ATCC_1997 AGHLIKTPGRILVVAGPEVARQDAREELADLVDRLDALVAVSPDAKDTVK
Mb3499c|M.bovis_AF2122/97 VIGRLDRSRAPMLVLGNGCRLDGIGEQIVAFCEKAGLPFATTPNGRGIVA
Rv3470c|M.tuberculosis_H37Rv VIGRLDRSRAPMLVLGNGCRLDGIGEQIVAFCEKAGLPFATTPNGRGIVA
MMAR_1710|M.marinum_M AAKLIGAARKPVLYVGGGVIRGEATEQLAELAELTGIPVVTTLMARGAFP
MUL_1947|M.ulcerans_Agy99 AAKLIGAARKPVLYVGGGVIRGEATEQLAELAELTGIPVVTTLMARGAFP
MLBr_01696|M.leprae_Br4923 AAKLIADARKPVLYVGGGVIRGEATEQLRDLAELTGIPVVSTLMARGAFP
Mflv_3445|M.gilvum_PYR-GCK AAEILNDGERVAILIGQGAR--GAHDEIAGVADVTGAGVAKALLGKDVLP
: * .: . .. .:.
MSMEG_1488|M.smegmatis_MC2_155 TPGFGSSSALGVCGVMGHPGVVAAAAGS--GACLLVGTRMTVTARAGLDG
Mvan_1352|M.vanbaalenii_PYR-1 -------SSIGVCGVMGHPDVADAVAAG--AACLLVGTRMAVTARAGLDA
TH_3455|M.thermoresistible__bu IPGLGSSSSLGVTGVMGHPSAAEAVAAS--AVCLLVGARLPVTARAGLDE
MAV_2365|M.avium_104 TPGFGLSSALGVTGIMGHPGVVEAVADS--ALCLVVGTRLAVTARAGLDD
MAB_2165|M.abscessus_ATCC_1997 SEN---PRLLGVIGVMGHPAVEQVMRHG--VTCVCVGTTMPVMTRGAVD-
Mb3499c|M.bovis_AF2122/97 ETH---PLSLGVLGIFGDGRADEYLFDTPCDLLIAVGVSFGGLVTRSFSP
Rv3470c|M.tuberculosis_H37Rv ETH---PLSLGVLGIFGDGRADEYLFDTPCDLLIAVGVSFGGLVTRSFSP
MMAR_1710|M.marinum_M DSH---RQNMGMPGMHGTVAAVAALQRS--DLLIALGTRFDDRVTGKLDS
MUL_1947|M.ulcerans_Agy99 DSH---RQNMGMPGMHGTVAAVAALQRS--DLLIALGSRFDDRVTGKLDS
MLBr_01696|M.leprae_Br4923 DSH---HQNLGMPGMHGTVAAVAALQRS--DLLIALGTRFDDRVTGKLDS
Mflv_3445|M.gilvum_PYR-GCK DDL---PFVTGSIGLLGTRPSYEMMRDC--DTLLIVGSNFPYTQFLPEFG
* *: * : :* :
MSMEG_1488|M.smegmatis_MC2_155 --VLGSVPTYSIGSQQPYVPATHVHTDDLRGALTMLAEALTGNGRP----
Mvan_1352|M.vanbaalenii_PYR-1 --ALAAVPTFSVGSGRPYVDCTHVPSGDLRQTLTVLARALADSPR-----
TH_3455|M.thermoresistible__bu --ALARVRTLSVGAVAPYVPCTHVHTEDLRTTLSQLTSALTGRGRP----
MAV_2365|M.avium_104 --ALAAVPTISLGSAPPYLACTHIHTDDLRASLRMLTSALTGRERP----
MAB_2165|M.abscessus_ATCC_1997 ---FSKIPVLSIGSAAPHVASRHLQTHDLVSTLAGLAAALP--ARP----
Mb3499c|M.bovis_AF2122/97 RWRGLKADVVHVDPDPSAVGRFVATSLGITTSGRAFVNALNCGR------
Rv3470c|M.tuberculosis_H37Rv RWRGLKADVVHVDPDPSAVGRFVATSLGITTSGRAFVNALNCGR------
MMAR_1710|M.marinum_M --FAPEAKVIHADIDPAEIGKNRNADVPIVGDVQAVITELIAMLRHYDIP
MUL_1947|M.ulcerans_Agy99 --FAPEAKVIHADIDPAEIGKNRNADVPIVGDVQAVITELIAMLRHYDIP
MLBr_01696|M.leprae_Br4923 --FAPDAKVIHADIDPAEIGKNRHADVPIVGDVKAVIVELIAMLRHYEVP
Mflv_3445|M.gilvum_PYR-GCK -----QARAVQIDDDGASIGMRYPTELNIVADAKATLAALQPHLRRR---
. . . : : *
MSMEG_1488|M.smegmatis_MC2_155 ----------HQIRVPDVVPHTELPTPPHDGPGIRYRDAMRVLDRALPDG
Mvan_1352|M.vanbaalenii_PYR-1 -------------TSPKALPHRELQPPPASGSGVRYRDAMRVLDAVLPAG
TH_3455|M.thermoresistible__bu ----------HGLRVPEISRRTELAPPPHDGPGVRYRDAMAALDAALPDG
MAV_2365|M.avium_104 ----------TGIRVPDAVSRTELNPPRFDGPGVRYREAMSMLDRVLPDG
MAB_2165|M.abscessus_ATCC_1997 ----------ETSDSETLYAPAHLEPPAHDGDGVRYHDVMHVLDRAIPAG
Mb3499c|M.bovis_AF2122/97 -----PPRFCRRVGVRPPAPAALPGTPQARGESIHPLELMHELDRELAPN
Rv3470c|M.tuberculosis_H37Rv -----PPRFCRRVGVRPPAPAALPGTPQARGESIHPLELMHELDRELAPN
MMAR_1710|M.marinum_M GNIEMSEWLAYLEGVRATYPLSYG--PQSDG-SLGPEYVIEKLGEIAGPD
MUL_1947|M.ulcerans_Agy99 GNIEMSEWLAYLEGVRAPYPLSYG--PQSDG-SLGPEYVIEKLGEIAGPD
MLBr_01696|M.leprae_Br4923 GNIEMTDWWSYLDGVRKTYPLSYS--PQSDG-TLSPEYVIEKLGEIVGPE
Mflv_3445|M.gilvum_PYR-GCK ---ADTAWRDTVEANVRRWWSVLEEQSTLSANPVNPMRVAWELSNRLPAN
. . : *.
MSMEG_1488|M.smegmatis_MC2_155 TDIVVDAGNIGASAIHHLPVRRDGRFLVALGMGGMGYSFGAGIGMTFHRA
Mvan_1352|M.vanbaalenii_PYR-1 TDIVVDAGNIGAAAIHHLPARRDGRFLVALGMGGMGYSFGAGIGMGFHR-
TH_3455|M.thermoresistible__bu ADIVIDAGNTGAAAIHYLPVRRGGRFAVALGMGGMGYSFGAAIGMAFGR-
MAV_2365|M.avium_104 SDIVVDVGSCGAAAVHYLPVRRGGRFVVAPGMG---YSFGAGIGMAVRRR
MAB_2165|M.abscessus_ATCC_1997 ADIFVDAGNTGAAAVHYLGARQHGRYVVALGMGGMGYAFGAGIGCAFAR-
Mb3499c|M.bovis_AF2122/97 ATICADVGTCISWTFRGIPVRRPGRFFATVDFSPMECGIAGAIGVALARP
Rv3470c|M.tuberculosis_H37Rv ATICADVGTCISWTFRGIPVRRPGRFFATVDFSPMGCGIAGAIGVALARP
MMAR_1710|M.marinum_M AIYVAGVGQHQMWAAQFIKYEKPRTWLNSGGLGTMGFAIPAAMGAKIALP
MUL_1947|M.ulcerans_Agy99 AIYVAGVGQHQMWAAQFIKYEKPRTWLNSGGLGTMGFAIPAAMGAKIALP
MLBr_01696|M.leprae_Br4923 AVYVAGVGQHQMWAAQFISYEKPRTWLNSGGLGTMGFAIPAAMGAKIARP
Mflv_3445|M.gilvum_PYR-GCK AIVTADSGSSTTWYARCVRFTEGVRGSVSGTLATMGCAVPYALGAKFAHP
: . * : : . : :. .. .:* .
MSMEG_1488|M.smegmatis_MC2_155 ARPGPHGQRRTVVIAGDGSFFMHG-MEIHTAVQYR-----LPITFLLFDN
Mvan_1352|M.vanbaalenii_PYR-1 -------RRPAVVIAGDGSFFMHG-TEIHTALQYR-----LPITFCLFDN
TH_3455|M.thermoresistible__bu -------RQRTVVIAGDGSFFMHG-LEIHTAIQYR-----LPITVVLFNN
MAV_2365|M.avium_104 K---FGRRGHVVVIAGDGAFFMHG-MEVHTAVQYR-----LPVAFVLINN
MAB_2165|M.abscessus_ATCC_1997 -------PGRTVVIAGDGAFFMHG-MEIHTAIEHR-----LPVTFVIVNN
Mb3499c|M.bovis_AF2122/97 -------EEHVICIAGDGAFLMHG-TEISTAVAHG-----IRVTWAVLND
Rv3470c|M.tuberculosis_H37Rv -------EEHVICIAGDGAFLMHG-TEISTAVAHG-----IRVTWAVLND
MMAR_1710|M.marinum_M -------DTEVWAIDGDGCFQMTN-QELATCAIEG-----IPIKVALINN
MUL_1947|M.ulcerans_Agy99 -------DTEVWAIDGDGCFQMTN-QELATCAIEG-----IPIKVALINN
MLBr_01696|M.leprae_Br4923 -------EAEVWAIDGDGCFQMTN-QELATCAIEG-----APIKVALINN
Mflv_3445|M.gilvum_PYR-GCK -------DRPVIALVGDGAMQMNGLAELLTVARYRREWDDLPFVICVFGN
. : ***.: * . *: * . :..:
MSMEG_1488|M.smegmatis_MC2_155 HAHAMCVTREQLFYGDRYTYNRFGPSH---LGAGLAAMLPGLTALDVDDI
Mvan_1352|M.vanbaalenii_PYR-1 HAHAMCVTREQLFYGDRYSYNRFGPSR---LGAGLAAMLPGLVAFDVTDV
TH_3455|M.thermoresistible__bu NAHAMCLTREQLFYDDRYSYNRFTRSN---LGAGLATMFPGLPAVDVADA
MAV_2365|M.avium_104 NAQAMCAMSEKLCYGNRHSYNTFAPSR---LGAGLAAMFPELPSVDVDDV
MAB_2165|M.abscessus_ATCC_1997 NAHAMCVTREQLYYGGSYSFNRFAPSH---IASGLGAMFPGLMCHSADTV
Mb3499c|M.bovis_AF2122/97 GQMSAS----AGPVSGRMDPSPVARIG--ANDLAAMARALGAEGIRVDTR
Rv3470c|M.tuberculosis_H37Rv GQMSAS----AGPVSGRMDPSPVARIG--ANDLAAMARALGAEGIRVDTR
MMAR_1710|M.marinum_M GNLGMVRQWQNLFYEERYSQTDLATHSHRIPDFVKLAEALGCVGLRCERE
MUL_1947|M.ulcerans_Agy99 GNLGMVRQWQNLFYEERYSQTDLATHSHRIPDFVKLAEALGCVGLRCERE
MLBr_01696|M.leprae_Br4923 GNLGMVRQWQALFYQERYSQTDLATHSHRIPDFVKLAEALGCVGLRCECE
Mflv_3445|M.gilvum_PYR-GCK GDLNQVTWELRAMGGSPKFEESQSLPA---VSYAEVARAMGVEARTVDSD
:
MSMEG_1488|M.smegmatis_MC2_155 DDLSAALDAALRTDGPVVISVECSPDEIPPFAAFLGT-ATETATATDPNT
Mvan_1352|M.vanbaalenii_PYR-1 DQLPAALMVALNQDGPAVVAVDCSADEIPPFAAFL--------------T
TH_3455|M.thermoresistible__bu ESLPQALAGALAADGPAVVSIECSADEIPPFAPFLAAPSAAADTQATPTT
MAV_2365|M.avium_104 DGFGAAVQSALEVDGPSVIGVECSAEEVPPVSAFLGP-VTPKSVECQHGM
MAB_2165|M.abscessus_ATCC_1997 ADLEAAMQQVMTHHGPSVVEVICSTDEIPPFAPFLSPEIAKERTHDRHDC
Mb3499c|M.bovis_AF2122/97 CELRAGVQKALAATG-PCVLDIAIDPEINK-PDIGLGR------------
Rv3470c|M.tuberculosis_H37Rv CELRAGVQKALAATG-PCVLDIAIDPEINK-PDIGLGR------------
MMAR_1710|M.marinum_M EDVADVINQAREISDRPVVIDFIVGADAQVWPMVAAGTSNDEIQAARGIR
MUL_1947|M.ulcerans_Agy99 EDVADVINQATEISDRPVVIDFIVGADAQVWPMVAAGTSNDEIQAARGIR
MLBr_01696|M.leprae_Br4923 EDVVDVINQARAINNRPVVIDFIVGADAQVWPMVAAGASNDEIQAARGIR
Mflv_3445|M.gilvum_PYR-GCK EDLGAAWDWALSATGPTLLDVRCDPEVPPIPPHVTFDQMLNMARAVAKGD
. . . : . .
MSMEG_1488|M.smegmatis_MC2_155 QENDSHATARA-----------
Mvan_1352|M.vanbaalenii_PYR-1 QKEESHADVHA-----------
TH_3455|M.thermoresistible__bu HEEKIDVAASA-----------
MAV_2365|M.avium_104 AQDRIDAIVGA-----------
MAB_2165|M.abscessus_ATCC_1997 KNGQSWPQSLARA---------
Mb3499c|M.bovis_AF2122/97 ----------------------
Rv3470c|M.tuberculosis_H37Rv ----------------------
MMAR_1710|M.marinum_M PLFDDESEGHA-----------
MUL_1947|M.ulcerans_Agy99 PLFDDESEGHA-----------
MLBr_01696|M.leprae_Br4923 PLFDDESEGHV-----------
Mflv_3445|M.gilvum_PYR-GCK PAARHLTYQTIKAKAAELFTRS