For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVATLFYTDELPDAGSLAVLGGDEGFHAATVRRIRPGERLVLGDGAGGLARCEVEHAGRDGLRARVLERW TVAPPNPPVTVVQALPKSERSELAVELATEAGADGFLAWQAARCVANWHGPRVDKGLRRWRAVAKAAARQ SRRPHIPTVDGVLSTASLTSRIRDEVAAGAVVLALHESATDRLTDVPVAQAKSLFLVIGPEGGIADDELA ALRQAGALSVRLGPQVLRTSTAAAVALGALGVLTPRWDDAASP
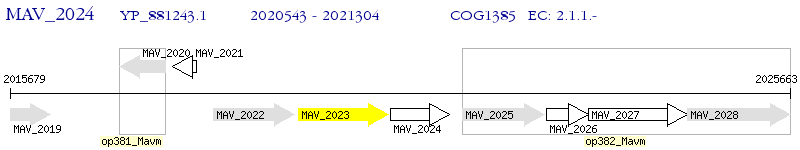
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2024 | - | - | 100% (253) | hypothetical protein MAV_2024 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2393c | - | 1e-108 | 77.38% (252) | 16S ribosomal RNA methyltransferase RsmE |
| M. gilvum PYR-GCK | Mflv_2703 | - | 1e-84 | 67.47% (249) | hypothetical protein Mflv_2703 |
| M. tuberculosis H37Rv | Rv2372c | - | 1e-108 | 77.38% (252) | 16S ribosomal RNA methyltransferase RsmE |
| M. leprae Br4923 | MLBr_00626 | - | 1e-108 | 77.38% (252) | 16S ribosomal RNA methyltransferase RsmE |
| M. abscessus ATCC 19977 | MAB_1667 | - | 4e-71 | 56.13% (253) | hypothetical protein MAB_1667 |
| M. marinum M | MMAR_3682 | - | 1e-109 | 79.76% (252) | hypothetical protein MMAR_3682 |
| M. smegmatis MC2 155 | MSMEG_4502 | - | 3e-88 | 66.94% (248) | hypothetical protein MSMEG_4502 |
| M. thermoresistible (build 8) | TH_1932 | - | 4e-88 | 68.03% (244) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3625 | - | 1e-109 | 79.05% (253) | 16S ribosomal RNA methyltransferase RsmE |
| M. vanbaalenii PYR-1 | Mvan_3835 | - | 6e-85 | 67.35% (245) | hypothetical protein Mvan_3835 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2024|M.avium_104 -MVATLFYTDELPDAGSLAVLGGDEGFHAATVRRIRPGERLVLGDGAGGL
MLBr_00626|M.leprae_Br4923 -MVSTLFYVDALPDTGTVAVVGGDEGFHAVTVRRIRPGEELVLGDGVGGL
Mb2393c|M.bovis_AF2122/97 -MVAMLFYVDTLPDTGAVAVVDGDEGFHAATVRRIRPGEQLVLGDGVGRL
Rv2372c|M.tuberculosis_H37Rv -MVAMLFYVDTLPDTGAVAVVDGDEGFHAATVRRIRPGEQLVLGDGVGRL
MMAR_3682|M.marinum_M -MVATLFYVDTVPEAGELAALDGDEGFHAATVRRIRPGEQLVLGDGAGEL
MUL_3625|M.ulcerans_Agy99 -MVATLFYVDTVPEAGELAALDGDEGFHAATVRRIRPGEQLVLGDGAGEL
MSMEG_4502|M.smegmatis_MC2_155 -MSAALFYVDVLPGAGELAVVDGDEGHHAANVRRIRVGERIDLSDGAGTL
Mflv_2703|M.gilvum_PYR-GCK --MRSLFYVDSLPGPGELAVVDGDEGHHAATVCRTRVGEHLDLSDGAGAV
Mvan_3835|M.vanbaalenii_PYR-1 --MRSLFYVDVVPAPGDLAVVDGEEGHHAATVCRTRPGEELDLADGAGTV
TH_1932|M.thermoresistible__bu -VSRALFYIDTVPAVGDTAVVDGDEGHHAANVRRIRVGEEIDLGDGAGVI
MAB_1667|M.abscessus_ATCC_1997 MAAATIFYVPRVPSAGEQVLLDGDEGHHAARVLRMRVGEPVMVCDGAGEV
:** :* * . :.*:**.**. * * * ** : : **.* :
MAV_2024|M.avium_104 ARCEVEHAGRDGLRARVLERWTVAPPNPPVTVVQALPKSERSELAVELAT
MLBr_00626|M.leprae_Br4923 AHCLVERAGRDGLYARVLGHWTVAPGLPPATVVQALPKAERSELAIEMAT
Mb2393c|M.bovis_AF2122/97 ARCVVEQAGRGGLRARVLRRWSVPPVRPPVTVVQALPKSERSELAIELAT
Rv2372c|M.tuberculosis_H37Rv ARCVVEQAGRGGLRARVLRRWSVPPVRPPVTVVQALPKSERSELAIELAT
MMAR_3682|M.marinum_M ARCVVEHAARDGLTARVLQRWSVAPVRPAVTVVQALPKSERSELAVELAT
MUL_3625|M.ulcerans_Agy99 ARCVVEHAARDGLTARVLQRWSVAPVRPAVTVVQALPKSERSELAVELAT
MSMEG_4502|M.smegmatis_MC2_155 AHCVVENTAKGSVSARITEQVTLPPARPSVTVVQALPKSDRSELAIELAT
Mflv_2703|M.gilvum_PYR-GCK AHCVVEDVAKGRLSARVQDRILVPVPTPAVTVVQALPKSDRSELAIELAT
Mvan_3835|M.vanbaalenii_PYR-1 AHCVVEDVAKGRLTARVQDRHLVAPPRPTVTVVQALPKSERSELAVELAT
TH_1932|M.thermoresistible__bu AHCVVEEAVRGRLAARVLDRRTVDLPTPTVTVVQALPKSDRAELAVETAT
MAB_1667|M.abscessus_ATCC_1997 GDGVVASVDRTGLAIEVTGRWSVAAVNPKVTVVQALPKSDRSDLAVDLAV
. * . : : .: : : * .********::*::**:: *.
MAV_2024|M.avium_104 EAGADGFLAWQAARCVANWHGP-RVDKGLRRWRAVAKAAARQSRRPHIPT
MLBr_00626|M.leprae_Br4923 EAGADAFVAWQSDRCVASWYGV-RVDKGLRRWRAVARAAARQSRRAHIPS
Mb2393c|M.bovis_AF2122/97 EAGADAFLAWQAARCVANWDGA-RVDKGLRRWRAVVRSAARQSRRARIPP
Rv2372c|M.tuberculosis_H37Rv EAGADAFLAWQAARCVANWDGA-RVDKGLRRWRAVVRSAARQSRRARIPP
MMAR_3682|M.marinum_M EAGADAFLAWQAARCVANWGGA-RADKGLRRWRSAARSAARQSRRAHIPP
MUL_3625|M.ulcerans_Agy99 EAGADAFLAWQAARCVANWGGA-RADKGLRRWRSAVRSAARQSRRAHIPP
MSMEG_4502|M.smegmatis_MC2_155 EAGADAFVAWQSARCVARWDGP-KADKGLRRWSAVARSAARQSRRAYIPT
Mflv_2703|M.gilvum_PYR-GCK EAGADAFLGWQAARCVARWDGAAKVDKGLRRWRAVARSAARQSRRPHIPE
Mvan_3835|M.vanbaalenii_PYR-1 EAGADAFIAWQASRCVARWDGAAKVDKGLRRWRAVARSAARQSRRPYIPD
TH_1932|M.thermoresistible__bu EAGADAFLAWQAARCVARWDGPQKVDKGLRRWRAVIRSAARQSRRAHIPA
MAB_1667|M.abscessus_ATCC_1997 EAGADEVIPWQAQRCVSRWDAH-KAEKGRRRWEAVAQAASRQSRRAHVPS
***** .: **: ***: * . :.:** *** :. ::*:*****. :*
MAV_2024|M.avium_104 VDGVLSTASLTSRIRDEVAAGAVVLALHESATDRLT----DVPVAQAKSL
MLBr_00626|M.leprae_Br4923 VDGVLTTAALLQRIRDEVAAGATVLALHESATDRLSDVFADISVAQANSL
Mb2393c|M.bovis_AF2122/97 VDGVLSTPMLVQRVREEVAAGAAVLVLHEEATERIV----DIAAAQAGSL
Rv2372c|M.tuberculosis_H37Rv VDGVLSTPMLVQRVREEVAAGAAVLVLHEEATERIV----DIAAAQAGSL
MMAR_3682|M.marinum_M VEGVLSTAALIRRITAEAQAGAMVLALHESATDRLV----DTAVAQAKSL
MUL_3625|M.ulcerans_Agy99 VEGVLSTAALIRRITAEAQAGAMVLALHESATDRLV----ETAVAQAKSL
MSMEG_4502|M.smegmatis_MC2_155 VTGVESTRDLLRRI---ASTDSVVLALHESATAPFA----EIPVAQSDSV
Mflv_2703|M.gilvum_PYR-GCK VRGPVSTQDLVGFTR--DAGPATVLVLHESATRPLT----DADLTAR-SL
Mvan_3835|M.vanbaalenii_PYR-1 VDGVVSTRDLVESTR--CAEPGTVLVLHESATRPLA----DAAAGQNDPL
TH_1932|M.thermoresistible__bu VGGVVSTAELTARVADDVAGGAVALVLHEAAAVPLT----AVGWQPVEAV
MAB_1667|M.abscessus_ATCC_1997 VPELHSTAQLAELVGARAAGGAVVLVLHESAESGLG--ALGARLSSASSV
* :* * . .*.*** * : .:
MAV_2024|M.avium_104 FLVIGPEGGIADDELAALRQAGALSVRLGPQVLRTSTAAAVALGALGVLT
MLBr_00626|M.leprae_Br4923 VLLVGPEGGIAPDEIAALTEAGAVAVRLGPTVLRSSTAAAVALGALGVLT
Mb2393c|M.bovis_AF2122/97 MLVVGPEGGIAPDELAALTDAGAVAVRLGPTVLRTSTAAAVALGAVGVLT
Rv2372c|M.tuberculosis_H37Rv MLVVGPEGGIAPDELAALTDAGAVAVRLGPTVLRTSTAAAVALGAVGVLT
MMAR_3682|M.marinum_M VLVVGPEGGIAPEEMDALAGAGAVSVRLGPTVLRTSTAAAVALGALGVLT
MUL_3625|M.ulcerans_Agy99 VLVVGPEGGIAPEEMDALAGAGAVSVRLGPTVLRTSTAAAVALGALGVLT
MSMEG_4502|M.smegmatis_MC2_155 ILIIGPEGGIADDEIAALTDAGAHAVRLGPTVLRTSTAAAVALGALGALT
Mflv_2703|M.gilvum_PYR-GCK TLVVGPEGGVADEELAALTDAGAMAVRLGPTVLRTSTAAAVALGALGVLT
Mvan_3835|M.vanbaalenii_PYR-1 TLIVGPEGGVADDELAALTDAGAVAVRLGPTVLRTSTAAAVALGALGVLT
TH_1932|M.thermoresistible__bu TLIVGPEGGITDDEIAELQEAGARAVRLGPTVLRTSTAAAVALGAIGVLT
MAB_1667|M.abscessus_ATCC_1997 VLIVGPEGGVADEELRILADAGAAPVRLGPTVLRTSSAAAVALGALGVLT
*::*****:: :*: * *** .***** ***:*:********:*.**
MAV_2024|M.avium_104 PRWDDAASP---------
MLBr_00626|M.leprae_Br4923 SRWDASSIL---------
Mb2393c|M.bovis_AF2122/97 SRWDASASDCEYCDVTRR
Rv2372c|M.tuberculosis_H37Rv SRWDASASDCEYCDVTRR
MMAR_3682|M.marinum_M ERWDSAASL---------
MUL_3625|M.ulcerans_Agy99 GRWDSAASP---------
MSMEG_4502|M.smegmatis_MC2_155 SRWNI-------------
Mflv_2703|M.gilvum_PYR-GCK GRWSDQTPRG--------
Mvan_3835|M.vanbaalenii_PYR-1 SRWS--------------
TH_1932|M.thermoresistible__bu PRWR--------------
MAB_1667|M.abscessus_ATCC_1997 DRWAAGPLVHR-------
**