For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSITPSLWFDDNLEEAATFYTAVFPNSHIEGFNRTTEAGPGEPGTVLSGSFVLDGSRFIGINGGPHFRF SEAVSFTVHCKDQDEVDYYWDRLSDGGEESQCGWLKDRFGLSWQIVPDRLFELIGDPDRSRAAAATQAMY GMRKIVIAELERAAASS
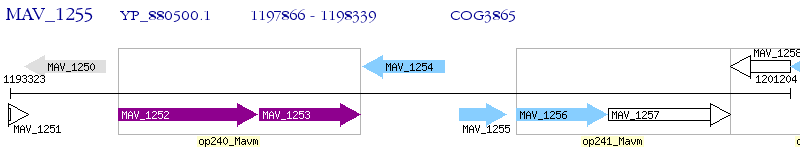
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1255 | - | - | 100% (157) | 3-demethylubiquinone-9 3-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2098 | - | 6e-60 | 67.10% (155) | 3-demethylubiquinone-9 3-methyltransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1279 | - | 7e-42 | 52.80% (161) | 3-demethylubiquinone-9 3-methyltransferase |
| M. marinum M | MMAR_4336 | - | 7e-73 | 81.70% (153) | hypothetical protein MMAR_4336 |
| M. smegmatis MC2 155 | MSMEG_0218 | - | 3e-27 | 40.65% (155) | 3-demethylubiquinone-9 3-methyltransferase |
| M. thermoresistible (build 8) | TH_2859 | - | 4e-71 | 77.56% (156) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0146 | - | 2e-72 | 81.70% (153) | hypothetical protein MUL_0146 |
| M. vanbaalenii PYR-1 | Mvan_4607 | - | 5e-62 | 70.59% (153) | 3-demethylubiquinone-9 3-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2098|M.gilvum_PYR-GCK ---------MP-----AITPSLWFDDNLEDAIGFYCSIFP-NSTHDSTEY
Mvan_4607|M.vanbaalenii_PYR-1 ---------MP-----TITPSLWFDNDLEEAIAYYSSIFP-DSAIETIER
MMAR_4336|M.marinum_M ---------MP-----AITPSLWFDHNLAEAAQFYTSVFP-NSQIEGFNR
MUL_0146|M.ulcerans_Agy99 ---------MP-----AITPSLWFDHNLAEAAQFYTSVFP-NSQIEGFNR
MAV_1255|M.avium_104 ---------MP-----SITPSLWFDDNLEEAATFYTAVFP-NSHIEGFNR
TH_2859|M.thermoresistible__bu ---------MP-----AIAPALWFDDNLEEAARFYTSIFP-NSSIEGFER
MAB_1279|M.abscessus_ATCC_1997 MVPAPVDTGMP-----TITPCLWFDTQAEEAANFYVSVFP-NSQILEVTH
MSMEG_0218|M.smegmatis_MC2_155 ---------MPVKINASIVPNLWFDREAEEAAEHYIAAFGGNGRILNKIA
** :*.* **** : :* .* : * :.
Mflv_2098|M.gilvum_PYR-GCK YTDAGPGTPGRALSATFTLDGQKFIGINGGPLFPFTEAVSFTIGCRDQTE
Mvan_4607|M.vanbaalenii_PYR-1 YNEAGPGTPGEVVAATFTLGGQRFIGINGGPQFPFTEAVSFTVACRDQAE
MMAR_4336|M.marinum_M HTEAGPGEPGTVISGTFVLDGVRFIGINGGPQFPFTEAVSFTVNCKDQDE
MUL_0146|M.ulcerans_Agy99 HTEAGPGEPGTVISGTFVLDGVRFIGINGGPPFPFTEAVSFTVNCKDQDE
MAV_1255|M.avium_104 TTEAGPGEPGTVLSGSFVLDGSRFIGINGGPHFRFSEAVSFTVHCKDQDE
TH_2859|M.thermoresistible__bu YTDAGPGTPGEVASGTFVLDGTRFVGINGGPAFSFTEAVSFMVTCADQDE
MAB_1279|M.abscessus_ATCC_1997 YGPNTPMPEGTVLTVEFELDGQRYTALNAGPQFTFDEAISFQIDCASQEE
MSMEG_0218|M.smegmatis_MC2_155 AHPDAPSPQDVPVVVEFELVGQRFVGINGGPQFTFNEAVSLEVRVAGQEQ
* . * * * :: .:*.** * * **:*: : .* :
Mflv_2098|M.gilvum_PYR-GCK VDYYWDRLVD-GGAASQCGWLKDRYGLSWQVVPDRLTELTSDPNPAIATA
Mvan_4607|M.vanbaalenii_PYR-1 VDYYWDRLVA-GGQESQCGWLKDRYGLSWQIVPDRLTELTSDPDPARAAA
MMAR_4336|M.marinum_M VDYYWDRLTQ-GGTESQCGWCKDRFGLSWQIVPDRLYELLSDPDPARAAA
MUL_0146|M.ulcerans_Agy99 VDYYWDRLTQ-GGTESQCGWCKDRFGLSWQIVPDRLHELLSDPDPARAAA
MAV_1255|M.avium_104 VDYYWDRLSD-GGEESQCGWLKDRFGLSWQIVPDRLFELIGDPDRSRAAA
TH_2859|M.thermoresistible__bu VDYYWDRLVD-GGQESQCGWLKDRFGLSWQIVPQRLFELVNDPDPARAAA
MAB_1279|M.abscessus_ATCC_1997 VDRYWAALTADGGQESQCGWLKDKFGLSWQVVSRELIDLIFHSDPVTGNA
MSMEG_0218|M.smegmatis_MC2_155 LDQVWAALTE-GGEELPCGWLKDKYGLAWQVTPSEYYDMVTKGDEEANNR
:* * * ** *** **::**:**:.. . :: . :
Mflv_2098|M.gilvum_PYR-GCK ATRAMLGMT-KIVVADLEEAVAGL----
Mvan_4607|M.vanbaalenii_PYR-1 ATKAMLGMR-KILIAQLEDAVQGV----
MMAR_4336|M.marinum_M ATTAMLGMH-KIVVAELEEAVNAAG---
MUL_0146|M.ulcerans_Agy99 ATTAMLGMH-KIVVAELEEALNAAG---
MAV_1255|M.avium_104 ATQAMYGMR-KIVIAELERAAASS----
TH_2859|M.thermoresistible__bu ATRAMLGMR-KIVIAELEEAVTNPV---
MAB_1279|M.abscessus_ATCC_1997 AMQAMMQMR-KLDVAVAREAVAKAQAGS
MSMEG_0218|M.smegmatis_MC2_155 LMTAVLSTHGKFDLAKLQAAYDNA----
*: *: :* . *