For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKFVSTRIITADVQRLVGFYEIVTRVSAVWANELFAEIPTPAATLAIGSDQTVPLFGAGSAEPAANRSAI VEFIVDDVDAEYERLREQLTEVVTEPTTMPWGNRALLFRDPDGNLVNLFTPVTPEARAKFGI
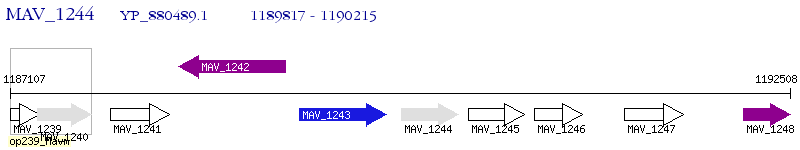
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1244 | - | - | 100% (132) | glyoxalase family protein |
| M. avium 104 | MAV_4932 | - | 5e-08 | 31.13% (106) | glyoxalase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4708c | - | 9e-06 | 40.00% (50) | putative glyoxalase/bleomycin resistance protein |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5216 | - | 3e-61 | 84.09% (132) | glyoxalase family protein |
| M. thermoresistible (build 8) | TH_0432 | - | 9e-05 | 26.09% (115) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3007 | - | 1e-06 | 42.00% (50) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1244|M.avium_104 ---------------------------MKFVSTRIITADVQRLVGFYEIV
MSMEG_5216|M.smegmatis_MC2_155 ---------------------------MKFVSTRIITADVKRLVDFYETV
MAB_4708c|M.abscessus_ATCC_199 MSSSRSPDRSGIEKQRLQCAASVTGMDITINASFLPHTDAEASLAFYRDA
Mvan_3007|M.vanbaalenii_PYR-1 -------------------------MDIMINASFLPHTDPEASLAFYRDA
TH_0432|M.thermoresistible__bu -------------------------MEILASRVLLRPADYQRTLAFYRDL
: : :* : : **.
MAV_1244|M.avium_104 TRVSA---VWANELFAEIPTPAATLAIGSDQTVPLFGAGSAEP-------
MSMEG_5216|M.smegmatis_MC2_155 TGLSA---VWANELFAEIPTPAGTLAIGSEKTVPLFGPGSAQP-------
MAB_4708c|M.abscessus_ATCC_199 LGFEVRKDVGYEGMRWITVGPAGRPDMNIVLTPPAADPGVTDEERRTITE
Mvan_3007|M.vanbaalenii_PYR-1 LGFEVRKDVGHQDMRWITLGPVGRPDTNIVLTPPAADPGVTDEERRVIAE
TH_0432|M.thermoresistible__bu IGLAI-----------AREYPGGTVFFAGQTLLEIAGHGGGEP-------
. * . . * :
MAV_1244|M.avium_104 --AANRSAIVEFIVDDVDAEYERLREQLTEVVTEPTTMPWGNRALLFRDP
MSMEG_5216|M.smegmatis_MC2_155 --AANHSAIIEFIVDDVDAEYERLRKHLVDVVIEPTTMPWGNRALVFRDP
MAB_4708c|M.abscessus_ATCC_199 MMAKGTYASIMLATDDLSGLFERVQAAGAEVMQEPTDQPWGARDCAFRDP
Mvan_3007|M.vanbaalenii_PYR-1 MMAKGTYASIMLATTDLDAVFERVQASGAEVMQEPTDQPWGARDCAFRDP
TH_0432|M.thermoresistible__bu --GGRFSGALWLQVRDVYAAQAELTERGVQITREARQERWGLHEMHVTDP
. . : : . *: . .: .:: *. ** : . **
MAV_1244|M.avium_104 DGNLVNLFTPVTPEARAKFGI----
MSMEG_5216|M.smegmatis_MC2_155 DGNLVNLFTPVTEDARAKFGVASA-
MAB_4708c|M.abscessus_ATCC_199 AGCTVRIQELA--------------
Mvan_3007|M.vanbaalenii_PYR-1 AGSTVRIQELP--------------
TH_0432|M.thermoresistible__bu DGVTLIFVQIPADHPLRRDTRMTED
* : :