For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSSRSPDRSGIEKQRLQCAASVTGMDITINASFLPHTDAEASLAFYRDALGFEVRKDVGYEGMRWITVGP AGRPDMNIVLTPPAADPGVTDEERRTITEMMAKGTYASIMLATDDLSGLFERVQAAGAEVMQEPTDQPWG ARDCAFRDPAGCTVRIQELA*
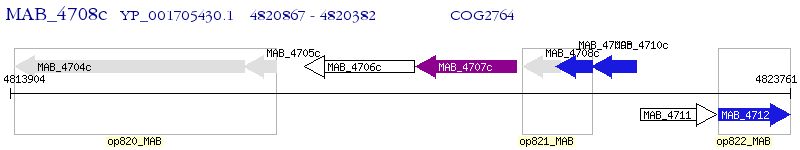
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4708c | - | - | 100% (161) | putative glyoxalase/bleomycin resistance protein |
| M. abscessus ATCC 19977 | MAB_3948 | - | 1e-06 | 26.67% (135) | hypothetical protein MAB_3948 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0560c | - | 7e-06 | 25.00% (140) | hypothetical protein Mb0560c |
| M. gilvum PYR-GCK | Mflv_2898 | - | 4e-22 | 39.13% (138) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0546c | - | 7e-06 | 25.00% (140) | hypothetical protein Rv0546c |
| M. leprae Br4923 | MLBr_02261 | - | 4e-05 | 23.70% (135) | hypothetical protein MLBr_02261 |
| M. marinum M | MMAR_5342 | - | 5e-08 | 29.10% (134) | hypothetical protein MMAR_5342 |
| M. avium 104 | MAV_1244 | - | 1e-05 | 40.00% (50) | glyoxalase family protein |
| M. smegmatis MC2 155 | MSMEG_0212 | - | 1e-48 | 65.93% (135) | lyase |
| M. thermoresistible (build 8) | TH_1881 | - | 3e-06 | 27.55% (98) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. ulcerans Agy99 | MUL_0087 | - | 3e-08 | 29.10% (134) | hypothetical protein MUL_0087 |
| M. vanbaalenii PYR-1 | Mvan_3007 | - | 1e-66 | 88.15% (135) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0560c|M.bovis_AF2122/97 -------------------------MEILASRMLLRPADYQRSLSFYRDQ
Rv0546c|M.tuberculosis_H37Rv -------------------------MEILASRMLLRPADYQRSLSFYRDQ
MLBr_02261|M.leprae_Br4923 -------------------------MEILASRMLLRPTDYQRSLSFYRDQ
MAB_4708c|M.abscessus_ATCC_199 MSSSRSPDRSGIEKQRLQCAASVTGMDITINASFLPHTDAEASLAFYRDA
Mvan_3007|M.vanbaalenii_PYR-1 -------------------------MDIMINASFLPHTDPEASLAFYRDA
MSMEG_0212|M.smegmatis_MC2_155 -----------------------MSNDLTIHNTFVPHEDPEESLAFYRDA
Mflv_2898|M.gilvum_PYR-GCK --------------------------MIKIASAHLWVHDQEVALKFWTQK
MMAR_5342|M.marinum_M ---------------------------MRINLASVLVDDQDKALRFYTEI
MUL_0087|M.ulcerans_Agy99 ---------------------------MRINLASVLVDDQDKALRFYTEI
TH_1881|M.thermoresistible__bu ----------------------------------VLVDDQDKALRLYTEK
MAV_1244|M.avium_104 ---------------------------MKFVSTRIITADVQRLVGFYEIV
: * : : ::
Mb0560c|M.bovis_AF2122/97 IGLAIAREYG-----AGTVFFAGQSLLELAGY---GEPDHSRGPFP----
Rv0546c|M.tuberculosis_H37Rv IGLAIAREYG-----AGTVFFAGQSLLELAGY---GEPDHSRGPFP----
MLBr_02261|M.leprae_Br4923 IGLAIAREYG-----TGTVFFAGQSLLELAGYVIQGAPDHSRGAFP----
MAB_4708c|M.abscessus_ATCC_199 LGFEVRKDV----GYEGMRWITVGPAGRPDMNIVLTPPAADPGVTDEERR
Mvan_3007|M.vanbaalenii_PYR-1 LGFEVRKDV----GHQDMRWITLGPVGRPDTNIVLTPPAADPGVTDEERR
MSMEG_0212|M.smegmatis_MC2_155 LGFEVRLDV----GHGTMRWITVGPPNQPGTSIVLYPPTADPAITEEEGK
Mflv_2898|M.gilvum_PYR-GCK VGMEVRQDVSLPDGDGVFRWLTVGPPGQDDVSIVLMAVPGEPLMDEPTRR
MMAR_5342|M.marinum_M LGFQTKHDVP----MGQFRWITVVSPEDPDGTELVLEPDEHPAAKP----
MUL_0087|M.ulcerans_Agy99 LGFQTKHDVP----MGQFRWITVVSPEDPDGTELVLEPDEHPAAKP----
TH_1881|M.thermoresistible__bu LGFLPKHDIP----LGDARWITVVSPEEPDGTELVLEPDGHPAVGP----
MAV_1244|M.avium_104 TRVSAVWANELF--AEIPTPAATLAIGSDQTVPLFGAGSAEPAANR----
. : .
Mb0560c|M.bovis_AF2122/97 -----------GALWLQVRDLEATQTELVSRGVSIAREPRREPWGLHEMH
Rv0546c|M.tuberculosis_H37Rv -----------GALWLQVRDLEATQTELVSRGVSIAREPRREPWGLHEMH
MLBr_02261|M.leprae_Br4923 -----------GALWLQVRDIAVTQADLEGRGVSITREPRREPWGLHEMH
MAB_4708c|M.abscessus_ATCC_199 TITEMMAKGTYASIMLATDDLSGLFERVQAAGAEVMQEPTDQPWGARDCA
Mvan_3007|M.vanbaalenii_PYR-1 VIAEMMAKGTYASIMLATTDLDAVFERVQASGAEVMQEPTDQPWGARDCA
MSMEG_0212|M.smegmatis_MC2_155 VIREMMAKGTFASIVLATADVDATFERLQATTAEIVQEPTDQPYGVRDCA
Mflv_2898|M.gilvum_PYR-GCK QVLDLTAKGFAGTIFLTTGDCHASYRELKARGVEFSEEPHDMPYGI-DCG
MMAR_5342|M.marinum_M -FKDALSSDGIPFTAFAVDNVQAEFDRLRALGVRFTQEPTEM-GPVTTAV
MUL_0087|M.ulcerans_Agy99 -FKDALSSDGIPFTAFAVDNVQAEFDRLRALGVRFTQEPTEM-GPVTTAV
TH_1881|M.thermoresistible__bu -FKEALVTDGIPFTSFAVDDVHAEFDRLRQMGVRFTQPPTTM-GNVITAV
MAV_1244|M.avium_104 ----------SAIVEFIVDDVDAEYERLREQLTEVVTEPTTMPWGNRALL
: . : : . . *
Mb0560c|M.bovis_AF2122/97 VTDPDGITLIFVEVPEGHPLRTDTRA-------
Rv0546c|M.tuberculosis_H37Rv VTDPDGITLIFVEVPEGHPLRTDTRA-------
MLBr_02261|M.leprae_Br4923 VTDPDEITLIFVEVPANHPLRRDTRSERPRTPD
MAB_4708c|M.abscessus_ATCC_199 FRDPAGCTVRIQELA------------------
Mvan_3007|M.vanbaalenii_PYR-1 FRDPAGSTVRIQELP------------------
MSMEG_0212|M.smegmatis_MC2_155 VRDPAGTMIRIQERAL-----------------
Mflv_2898|M.gilvum_PYR-GCK FRDPSGNSVRLTQLADVPAFG------------
MMAR_5342|M.marinum_M LDDTCGNLIQIQQTSSLLAGDGSGGR-------
MUL_0087|M.ulcerans_Agy99 LDDTCGNLIQIQQAGSQPASEGSGG--------
TH_1881|M.thermoresistible__bu LDDTCGNLIQLAQMIG-----------------
MAV_1244|M.avium_104 FRDPDGNLVNLFTPVTPEARAKFGI--------
. *. : :