For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDPQPPRNEFGVASVLLGLVGLVTCWLLLGVPFGIAAVVTGDIARRRVARGEASNPRTAMTGIVLGAVAI LAGLVAISYYARMDAQNR*
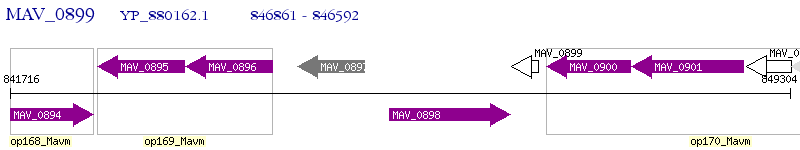
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0899 | - | - | 100% (89) | hypothetical protein MAV_0899 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3589 | - | 1e-11 | 44.00% (75) | hypothetical protein Mflv_3589 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2634 | - | 1e-07 | 37.14% (70) | hypothetical protein MAB_2634 |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3229 | - | 5e-13 | 39.24% (79) | hypothetical protein MSMEG_3229 |
| M. thermoresistible (build 8) | TH_2549 | - | 1e-14 | 43.90% (82) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2826 | - | 1e-18 | 52.38% (84) | hypothetical protein Mvan_2826 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2634|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3229|M.smegmatis_MC2_155 MFAVPAVSRWREGSLTDMTDERAGEPGSDRPEPTESSGQPSDPGQTPPPT
TH_2549|M.thermoresistible__bu --------------------------------------------------
Mflv_3589|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2826|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_0899|M.avium_104 --------------------------------------------------
MAB_2634|M.abscessus_ATCC_1997 -------MTETPPPPP---------------------PY-----------
MSMEG_3229|M.smegmatis_MC2_155 RSGESTESSDTASPPPESNPPYTYGASQGAYDTASQAPYGTAPQAPYGSQ
TH_2549|M.thermoresistible__bu -MTEPDKPSVPEQPQPN--------------------PYG----------
Mflv_3589|M.gilvum_PYR-GCK ----MSEPPLPPPPPPG--------------------TYPGG--------
Mvan_2826|M.vanbaalenii_PYR-1 ----MSEP---PPPPPG--------------------SYPGG--------
MAV_0899|M.avium_104 --------------------------------------------------
MAB_2634|M.abscessus_ATCC_1997 ----QPAGGYPPPFP--YPYPYGAP-GVPVPPPAP-----KNGLGLSALV
MSMEG_3229|M.smegmatis_MC2_155 PYGSQPYGAYPGAYPGSYPPPYGG--GYPAQPPAQGAAGPRNGLGIAALI
TH_2549|M.thermoresistible__bu ----QPPGGYPVPPP---PPPYGGYPGHPAAPTAP-----QNGLGIAALV
Mflv_3589|M.gilvum_PYR-GCK -YPPQPYGGYPGPYS---PQPY---------PVAP-----KNGLGTASLV
Mvan_2826|M.vanbaalenii_PYR-1 -YPPQPYGAYPPAYP---TQPYGGY--PPSPPTAP-----KNGLGTAALV
MAV_0899|M.avium_104 ---------MSDPQP-------------------P-----RNEFGVASVL
. . . :* :* ::::
MAB_2634|M.abscessus_ATCC_1997 IGIVSVLLSCTVFGGLFGGLVAVVLGIAGLLRVKRAVADNRGVAIAGIAL
MSMEG_3229|M.smegmatis_MC2_155 AAIAGLVLCWSIIGGLIVGVVAIVLGFLGHGRAKRGEATNGGVAIAGIVL
TH_2549|M.thermoresistible__bu VAIVALLFFWSVLGGIVGGIVAVILGVLARGRVKRGEATNGGVATAGIVL
Mflv_3589|M.gilvum_PYR-GCK VAIIS---MFTLYGGVILGVVAVVLGFLGRGRVKRGEANNGGVALAGIVL
Mvan_2826|M.vanbaalenii_PYR-1 LAIVGLLLCWTVAGGVLLGIAAIVMGFVARGRVKRGEATNGGVALAGIVL
MAV_0899|M.avium_104 LGLVGLVTCWLLLG-VPFGIAAVVTGDIARRRVARGEASNPRTAMTGIVL
.: . : * : *:.*:: * . *. *. * * .* :**.*
MAB_2634|M.abscessus_ATCC_1997 GSLAVLLS-ATILVFT---LVFMKSAGLNDFISCLSDVKGDQAKATQCQN
MSMEG_3229|M.smegmatis_MC2_155 GGIAVVLSLAFIAIWATLGFKWFEELGGREYIACLEDAGNDQAAQERCRV
TH_2549|M.thermoresistible__bu GVLAVLAGVASIAIYV----ALFNEVGGADYVSCVRDAGNDDAALQECEQ
Mflv_3589|M.gilvum_PYR-GCK GALSIVVAIAAIIITAIFGWQLFQDVGGNDYIDCLNRAGSDQAAVEQCAN
Mvan_2826|M.vanbaalenii_PYR-1 GAVAIVASLVIVAIYATLGVTLFKDLGGGDYLDCLQRAGNDQQAVQECAD
MAV_0899|M.avium_104 GAVAILAGLVAISYYA----------------------------------
* :::: . . : .
MAB_2634|M.abscessus_ATCC_1997 DFEKRMNEKAGTSKP---
MSMEG_3229|M.smegmatis_MC2_155 QFEERVEDEFGTPQSR--
TH_2549|M.thermoresistible__bu RFRDRLEDRLGVTLEPNP
Mflv_3589|M.gilvum_PYR-GCK EFTQRVENEFSITVTPTP
Mvan_2826|M.vanbaalenii_PYR-1 EFTQRVEESFSITVTPTP
MAV_0899|M.avium_104 ----RMDAQNR-------
*::