For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGSGTGSQTLARGLTALQMVADAPAGLTVQQLADQVGVHRTIAYRLLTTLAEFRLVAKGEDGRYRPAAGL AVLGASFDRNVRQLCLPTLRALADELGTTVSLLVAEGDQQVAIAVIVPSHVAYQLSFHEGSRYPLDRGAA GIALLACMPPRPGERELVSRARERGWVTTYGEIEPNTYGLAVGVRRPAPSPPTCINLISHREDVVMRGKD AVMRAAEQLSKLLS*
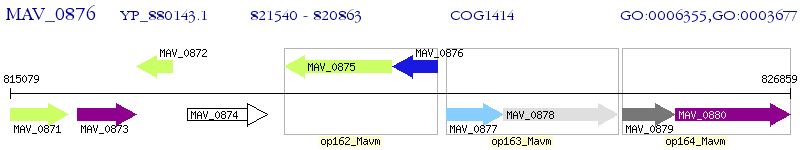
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0876 | - | - | 100% (225) | transcriptional regulator |
| M. avium 104 | MAV_3839 | - | 2e-12 | 28.09% (178) | transcriptional regulator, IclR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3013 | - | 1e-14 | 26.84% (190) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_2399 | - | 2e-96 | 79.00% (219) | regulatory protein, IclR |
| M. tuberculosis H37Rv | Rv2989 | - | 1e-14 | 26.84% (190) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3295 | - | 1e-12 | 28.27% (191) | IclR family transcriptional regulator |
| M. marinum M | MMAR_4779 | - | 1e-102 | 82.67% (225) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_4868 | - | 3e-99 | 81.65% (218) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_0967 | - | 2e-93 | 77.52% (218) | transcriptional regulator, IclR family protein, C- domain |
| M. ulcerans Agy99 | MUL_0348 | - | 1e-102 | 82.67% (225) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4247 | - | 8e-98 | 78.57% (224) | regulatory proteins, IclR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4779|M.marinum_M -------------------MTGSGAGSQTLARGLTALQLVAASPAGLSMQ
MUL_0348|M.ulcerans_Agy99 -------------------MTGSGAGSQTLARGLTALQLVAASPAGLSMQ
MAV_0876|M.avium_104 -------------------MTGSGTGSQTLARGLTALQMVADAPAGLTVQ
MSMEG_4868|M.smegmatis_MC2_155 ---MPANAAQSAARTRVEGAPSAAPGSQTLARGLSALQAIADAPGGLTVQ
TH_0967|M.thermoresistible__bu ---------MTSVKGRGAPAERPAAVSQTLARGLSALQAVAGAPDGLTVQ
Mflv_2399|M.gilvum_PYR-GCK -----------MADPGNANAGPGAPGSQTLARGLNALQLVAGSPNGLTVA
Mvan_4247|M.vanbaalenii_PYR-1 MNRPAGDQAPVTGEPGPATAGAGAPGSQTLARGLNALQLVAKSPTGLTVG
Mb3013|M.bovis_AF2122/97 --------------------MRQHSGIGVLDKAVGVLHAVAESPCGL--A
Rv2989|M.tuberculosis_H37Rv --------------------MRQHSGIGVLDKAVGVLHAVAESPCGL--A
MAB_3295|M.abscessus_ATCC_1997 --------------------MRQHSGIGVLDKAVQVLTAIGDSPCGL--A
. .* :.: .* :. :* **
MMAR_4779|M.marinum_M EVADRVGVHRTIAYRLLLTLTQFRLVAKGEDGRFRPAAGLAVLGASFDHN
MUL_0348|M.ulcerans_Agy99 EVADRVGVHRTIAYRLLLTLTQFRLVAKGEDGRFRPAAGLAVLGASFDHN
MAV_0876|M.avium_104 QLADQVGVHRTIAYRLLTTLAEFRLVAKGEDGRYRPAAGLAVLGASFDRN
MSMEG_4868|M.smegmatis_MC2_155 QVADHLGAHRTIAYRLLATLSQFRYVAKGEDGRYRPAAALAVLGSSFDNN
TH_0967|M.thermoresistible__bu QVADEVGVHRTIAYRLLGTLSQFRYVIRGDDGRYRPAAALALLGAAFDNN
Mflv_2399|M.gilvum_PYR-GCK QVAEDIGVHRTIAYRLLSTLAQYRFVSKGEDGRYRSAAALAVLGASFDNN
Mvan_4247|M.vanbaalenii_PYR-1 QVAEDIGVHRTIAYRLLSTLAQYRFVAKGEDGRYRSAAALAVLGASFDNN
Mb3013|M.bovis_AF2122/97 ELCDRTDLPRATAYRLAAALEVHRLLGRGQDGHWRLGPAITELATHVDDP
Rv2989|M.tuberculosis_H37Rv ELCDRTDLPRATAYRLAAALEVHRLLGRGQDGHWRLGPAITELATHVDDP
MAB_3295|M.abscessus_ATCC_1997 ELCERTGLPRATAHRLAAGLEVHRLLSRDGDGQWQLGAGLTELAAHVNDP
::.: . *: *:** * .* : :. **::: ...:: *.: .:
MMAR_4779|M.marinum_M VRQLSLPTLRALADDLGTTVSLLVAEGDEQVALAVIVPTQVAYQLAFHEG
MUL_0348|M.ulcerans_Agy99 VRQLSLPTLRALADDLGTTVSLLVAEGDEQVALAVIVPTQVAYQLAFHEG
MAV_0876|M.avium_104 VRQLCLPTLRALADELGTTVSLLVAEGDQQVAIAVIVPSHVAYQLSFHEG
MSMEG_4868|M.smegmatis_MC2_155 VRQLAIPTLRTLADELGTTVSLLVAEGDQQVAIAVIVPSQVYYQLSFHEG
TH_0967|M.thermoresistible__bu LRQYSVPVLRALADEVGTTVSLLVAEGDQQVAIAVIVPTQVFYQLSFHEG
Mflv_2399|M.gilvum_PYR-GCK VRQLCVPTLRGLADELGTTVSLLVAEGDQQVAIAVMVPTNVFYQLSFHEG
Mvan_4247|M.vanbaalenii_PYR-1 VRQLCVPTLRALADELGTTVSLLVAEGDQQVAIAVMVPTNVFYQLSFHEG
Mb3013|M.bovis_AF2122/97 LLVACAAVLPQLRDATGESVQVYRREGTSRVCVAALEP-AAGLRDTVPVG
Rv2989|M.tuberculosis_H37Rv LLVACAAVLPQLRDATGESVQVYRREGTSRVCVAALEP-AAGLRDTVPVG
MAB_3295|M.abscessus_ATCC_1997 LLSASAQVLPKLREITGESVQLYRREGTVRICVAALEP-PAGLRDTVPVG
: . .* * : * :*.: ** ::.:*.: * . : :. *
MMAR_4779|M.marinum_M SRYPLDRGAAGIALLAS---------MAARPGERELVAATRRRGWVTTFG
MUL_0348|M.ulcerans_Agy99 SRYPLDRGAAGIALLAS---------MAARPGERELVAATRRRGWVTTFG
MAV_0876|M.avium_104 SRYPLDRGAAGIALLAC---------MPPRPGERELVSRARERGWVTTYG
MSMEG_4868|M.smegmatis_MC2_155 SRHPLDRGAAGVALLAS---------MPPRPGERDLVRQTREQGWVITHG
TH_0967|M.thermoresistible__bu SRYPIDRGAAGIALLAS---------MPPRPGERDLVRQTREQGWVITHG
Mflv_2399|M.gilvum_PYR-GCK SRYPLDRGAAGIALLAS---------MPPRPQERDLVRQARQQGWVITHG
Mvan_4247|M.vanbaalenii_PYR-1 SRYPLDRGAAGIALLAS---------MPPRPGERDLVQQTRQQGWVITHG
Mb3013|M.bovis_AF2122/97 ARLPMTAGSGAKVLLAHTDAATQAAVLPKAVFSARALAEVCRRGWAQSVA
Rv2989|M.tuberculosis_H37Rv ARLPMTAGSGAKVLLAHTDAATQAAVLPKAVFSARALAEVCRRGWAQSVA
MAB_3295|M.abscessus_ATCC_1997 SRLPMSAGSGAKVLLAHSDPGTQAAVLPGATFTERALADVRKRGWAQSAA
:* *: *:.. .*** :. : . .:**. : .
MMAR_4779|M.marinum_M EIEPNTYGLAVPVSRPAPAPPTCINLISHREDVVMRA----KGAVIDAAR
MUL_0348|M.ulcerans_Agy99 EIEPNTYGLAVPVSRPAPAPPTCINLISHREDVVMRA----KGAVIDAAR
MAV_0876|M.avium_104 EIEPNTYGLAVGVRRPAPSPPTCINLISHREDVVMRG----KDAVMRAAE
MSMEG_4868|M.smegmatis_MC2_155 EIEPNTYGLAVPVRRRPPSPPTCINLISHREDVVLGG----KDAVVRAAR
TH_0967|M.thermoresistible__bu EIEPDTYGLAVPVRRRPHAPATCINLISHREDVVRRG----KDAVVRAAQ
Mflv_2399|M.gilvum_PYR-GCK EIEPNTFGLAVPVRRRQPSPPTCINLISHREDVVIDG----RDAVIKAAN
Mvan_4247|M.vanbaalenii_PYR-1 EIEPNTYGLAVPVRRRPPSPPTCINLISHREDVVLDG----RDSVIKAAA
Mb3013|M.bovis_AF2122/97 EREPGVASVSAPVRDGRGVVIAAISVSGPIDRMGRRPGVRWAADLLSAAD
Rv2989|M.tuberculosis_H37Rv EREPGVASVSAPVRDGRGVVIAAISVSGPIDRMGRRPGVRWAADLLSAAD
MAB_3295|M.abscessus_ATCC_1997 EREAGVASVSAPVRDRHGNVIAAISVSGPIDRMGRRPGARWAADLVAASE
* *... .::. * :.*.: . : : :: *:
MMAR_4779|M.marinum_M QLSTQLS
MUL_0348|M.ulcerans_Agy99 QLSTQLS
MAV_0876|M.avium_104 QLSKLLS
MSMEG_4868|M.smegmatis_MC2_155 ELSAILH
TH_0967|M.thermoresistible__bu ELSAILN
Mflv_2399|M.gilvum_PYR-GCK ELSAVLS
Mvan_4247|M.vanbaalenii_PYR-1 ELSEALN
Mb3013|M.bovis_AF2122/97 ALTRRL-
Rv2989|M.tuberculosis_H37Rv ALTRRL-
MAB_3295|M.abscessus_ATCC_1997 AITARL-
:: *