For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADIMPQPPAVGSPLISVTDLLEWYRVARRDLPWRAPGVSAWQILVSEFMLQQTPVSRVLPIWPDWVRRWP TPSATAAASAADVLRAWGKLGYPRRAKRLHECATVIARDHGDVVPRDIDTLLTLPGVGGYTARAVACFAY RRPVPVVDTNVRRVVARAVHGQADAGAPSAGRDHADVAALLPGDGSAPEFSVALMELGATVCTARAPRCG LCPLRRCAWREAGHPPPTGPARRVQTYAGTDRQVRGRLMDVLRGNDSPVTRAELDVAWLTDTAQRDRALY SLLADGLVTQTADGRFALAGEE*
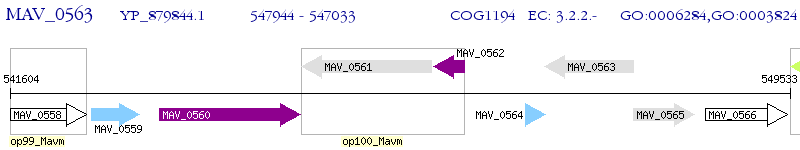
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0563 | - | - | 100% (303) | putative A/G-specific adenine glycosylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3620 | mutY | 1e-140 | 79.80% (302) | putative adenine glycosylase |
| M. gilvum PYR-GCK | Mflv_1438 | - | 1e-134 | 81.25% (288) | HhH-GPD family protein |
| M. tuberculosis H37Rv | Rv3589 | mutY | 1e-140 | 79.80% (302) | adenine glycosylase MutY |
| M. leprae Br4923 | MLBr_01920 | mutY | 1e-135 | 76.49% (302) | putative DNA glycosylase |
| M. abscessus ATCC 19977 | MAB_0558c | - | 1e-114 | 71.89% (281) | adenine glycosylase MutY |
| M. marinum M | MMAR_5089 | mutY | 1e-141 | 83.97% (287) | adenine glycosylase MutY |
| M. smegmatis MC2 155 | MSMEG_6083 | - | 1e-124 | 76.66% (287) | base excision DNA repair protein, HhH-GPD family protein |
| M. thermoresistible (build 8) | TH_0591 | mutY | 1e-123 | 75.70% (284) | PROBABLE ADENINE GLYCOSYLASE MUTY |
| M. ulcerans Agy99 | MUL_4165 | mutY | 1e-144 | 81.85% (303) | adenine glycosylase MutY |
| M. vanbaalenii PYR-1 | Mvan_5346 | - | 1e-133 | 79.51% (288) | HhH-GPD family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1438|M.gilvum_PYR-GCK --------------MIDPNELIRWYGTAQRDLAWRRPGVSAWQILVSEFM
Mvan_5346|M.vanbaalenii_PYR-1 --------------MIDPDALLDWYEHAQRDLPWRRPGVSAWQILVSEFM
Mb3620|M.bovis_AF2122/97 MPHILPEPSVTGPRHISDTNLLAWYQRSHRDLPWREPGVSPWQILVSEFM
Rv3589|M.tuberculosis_H37Rv MPHILPEPSVTGPRHISDTNLLAWYQRSHRDLPWREPGVSPWQILVSEFM
MMAR_5089|M.marinum_M ---------MTCPEHIPATNLVAWYERCCRDLPWRAPDVSPWQILVSEFM
MUL_4165|M.ulcerans_Agy99 MPAIVPQPSMTCPEHIPATNLVAWYERCYRDLPWRAPDVSPWQILVSEFM
MAV_0563|M.avium_104 MADIMPQPPAVGSPLISVTDLLEWYRVARRDLPWRAPGVSAWQILVSEFM
MLBr_01920|M.leprae_Br4923 MAHIVPQ------IKISTGGLLDWYRQSCRDLPWRQPGVTAWQILVSEFM
MSMEG_6083|M.smegmatis_MC2_155 -------------MSISPVELLSWYDHARRDLPWRRPGVSAWQILVSEFM
TH_0591|M.thermoresistible__bu ---------------MRATELLGWYEHARRDLPWRRPGVTPWQILVSEVM
MAB_0558c|M.abscessus_ATCC_199 ---------------------MGWFDVAERDLPWRRPEATPWHILISEVM
: *: . ***.** * .:.*:**:**.*
Mflv_1438|M.gilvum_PYR-GCK LQQTPVARVEPIWLDWVSRWPTPSATAAASAADILRAWGKLGYPRRAKRL
Mvan_5346|M.vanbaalenii_PYR-1 LQQTPVARVEPIWLDWIARWPTPSATAAAGPADILRAWGKLGYPRRAKRL
Mb3620|M.bovis_AF2122/97 LQQTPAARVLAIWPDWVRRWPTPSATATASTADVLRAWGKLGYPRRAKRL
Rv3589|M.tuberculosis_H37Rv LQQTPAARVLAIWPDWVRRWPTPSATATASTADVLRAWGKLGYPRRAKRL
MMAR_5089|M.marinum_M LQQTPVSRVLSIWPDWVRRWPTASATATATAADVLRAWGKLGYPRRAKRL
MUL_4165|M.ulcerans_Agy99 LQQTPVSRVLSIWPDWVRRWPTASATATATAADVLRAWGKLGYPRRAKRL
MAV_0563|M.avium_104 LQQTPVSRVLPIWPDWVRRWPTPSATAAASAADVLRAWGKLGYPRRAKRL
MLBr_01920|M.leprae_Br4923 LQQTPVARVLSIWPDWVQRWPTPSTTAAASAADVLRAWGKLGYPRRAKRL
MSMEG_6083|M.smegmatis_MC2_155 LQQTPVSRVEPIWSAWIERWPTASATAAAGPAEVLRAWGKLGYPRRAKRL
TH_0591|M.thermoresistible__bu LQQTPVARVAPVWTEWVTRWPTPSATAAAGVADVLRAWGRLGYPRRAKRL
MAB_0558c|M.abscessus_ATCC_199 LQQTPVSRVEPVWREWVARWPVPSAMAKTSVAEVLRAWGKLGYPRRAMRL
*****.:** .:* *: ***..*: * : *::*****:******* **
Mflv_1438|M.gilvum_PYR-GCK HECATVIATEHGDVVPDDVDTLVTLPGVGTYTARAVACFAYRQRVPVVDT
Mvan_5346|M.vanbaalenii_PYR-1 HECATVIATDHDDTVPDDVETLLTLPGIGTYTARAVACFAYRQRVPVVDT
Mb3620|M.bovis_AF2122/97 HECATVIARDHNDVVPDDIEILVTLPGVGSYTARAVACFAYRQRVPVVDT
Rv3589|M.tuberculosis_H37Rv HECATVIARDHNDVVPDDIEILVTLPGVGSYTARAVACFAYRQRVPVVDT
MMAR_5089|M.marinum_M HECAMVVAGDHDDVVPDDVDTLLTLPGVGSYTARAVACFAYRQRVPVVDT
MUL_4165|M.ulcerans_Agy99 HECAMVIAGDHDDVVPDDVDTLLTLPGVGSYTARAVACFAYRQRVPVVDT
MAV_0563|M.avium_104 HECATVIARDHGDVVPRDIDTLLTLPGVGGYTARAVACFAYRRPVPVVDT
MLBr_01920|M.leprae_Br4923 HESATIIAREHDDVVPDDVDVLLTMPGIGSYTARAVACFAYHQPVPVVDT
MSMEG_6083|M.smegmatis_MC2_155 HECAVVIASEYDDVVPRDVDTLLTLPGIGAYTARAVACFAYQASVPVVDT
TH_0591|M.thermoresistible__bu HECAVIIATRFDDVVPSDVDTLLTLPGVGSYTARAVACFAYRQRVPVVDT
MAB_0558c|M.abscessus_ATCC_199 HECATVLARDYDDQVPGDVETLLTLPGVGAYTARAIACFGYGQRVPVVDT
**.* ::* ..* ** *:: *:*:**:* *****:***.* ******
Mflv_1438|M.gilvum_PYR-GCK NVRRVVARAVHGLHDAGPPST-RDLADVAALLPDDDTAPHFSIAVMELGA
Mvan_5346|M.vanbaalenii_PYR-1 NVRRVVARAVHGLADAGSPSS-RDLGDVETLLPADDTAPRFSVALMEFGA
Mb3620|M.bovis_AF2122/97 NVRRVVARAVHGRADAGAPSVPRDHADVLALLPHRETAPEFSVALMELGA
Rv3589|M.tuberculosis_H37Rv NVRRVVARAVHGRADAGAPSVPRDHADVLALLPHRETAPEFSVALMELGA
MMAR_5089|M.marinum_M NVRRVVARAVHGRAEAGSPSATRDHAEVSALLPDDELAPRFSVALMELGA
MUL_4165|M.ulcerans_Agy99 NVRRVVARAVHGRAEAGSPSATRDHAEVSALLPDDELAPRFSVALMELGA
MAV_0563|M.avium_104 NVRRVVARAVHGQADAGAPSAGRDHADVAALLPGDGSAPEFSVALMELGA
MLBr_01920|M.leprae_Br4923 NVRRVVARAVHGRADAGPPSATRDHADVSSLLPGKKAAPQFSMALMELGA
MSMEG_6083|M.smegmatis_MC2_155 NVRRVVTRAVHGAADA--PASTRDLDMVAALLPPDTTAPTFSAALMELGA
TH_0591|M.thermoresistible__bu NVRRVVTRVVHGVAHA--PPSSRDLDEVAALLPDDDTAPRFSAALMELGA
MAB_0558c|M.abscessus_ATCC_199 NVRRVIARVVHGVADS--APSARDLRDAEALLPTEN-GARFSAALMELGA
*****::*.*** .: .. ** . :*** .. ** *:**:**
Mflv_1438|M.gilvum_PYR-GCK TVCTARAPRCGVCPLTHCAWRSRGFPAGTGPARRVQRYAGTDRQVRGRLL
Mvan_5346|M.vanbaalenii_PYR-1 TVCTARAPRCGVCPLQRCGWRARGFPAGTGPARRAQKYAGTDRQARGRLL
Mb3620|M.bovis_AF2122/97 TVCTARTPRCGLCPLDWCAWRHAGYPPSDGPPRRGQAYTGTDRQVRGRLL
Rv3589|M.tuberculosis_H37Rv TVCTARTPRCGLCPLDWCAWRHAGYPPSDGPPRRGQAYTGTDRQVRGRLL
MMAR_5089|M.marinum_M TVCTARAPRCGQCPLAECAWRRAGYPPATGPARRVQTYAGTDRQVRGRLL
MUL_4165|M.ulcerans_Agy99 TVCTARAPRCGQCPLAECAWRRAGYPPATGPARRVQTYAGTDRQVRGRLL
MAV_0563|M.avium_104 TVCTARAPRCGLCPLRRCAWREAGHPPPTGPARRVQTYAGTDRQVRGRLM
MLBr_01920|M.leprae_Br4923 IVCTARSPRCGLCPLNECAWRHAGYPPAQGPARRAQQYAGTDRQVRGLLL
MSMEG_6083|M.smegmatis_MC2_155 TVCTARSPRCGICPLSHCRWRSAGFPAGT-VARRVQRYAGTDRQVRGKLL
TH_0591|M.thermoresistible__bu TVCTARSPRCGDCPLSRCRWRERKYPPAT-APRRVQRYAGTDRQVRGLLL
MAB_0558c|M.abscessus_ATCC_199 LVCTARTPQCPMCPLSSCAWHEAGQPPLDAPARPVQKYAGTDRQVRGRLL
*****:*:* *** * *: *. .* * *:*****.** *:
Mflv_1438|M.gilvum_PYR-GCK DVLRDNASPVTRAELDVAWTTDPAQRDRALDSLLVDGLVEQTADGRFALA
Mvan_5346|M.vanbaalenii_PYR-1 DVLRGSDSPVTRAELDVAWTTDTAQRDRALDSLLVDGLVEQTADGRFALC
Mb3620|M.bovis_AF2122/97 DVLRAAEFPVTRAELDVAWLTDTAQRDRALESLLADALVTRTVDGRFALP
Rv3589|M.tuberculosis_H37Rv DVLRAAEFPVTRAELDVAWLTDTAQRDRALESLLADALVTRTVDGRFALP
MMAR_5089|M.marinum_M DVLRDNNGPVTRAQLDVAWLTDTAQRDRALYSLLADGLVTQTADGLFALA
MUL_4165|M.ulcerans_Agy99 DVLRDNNGPVTRAQLDVAWLTDTAQRDRALYSLLADGLVTQTADGLFALA
MAV_0563|M.avium_104 DVLRGNDSPVTRAELDVAWLTDTAQRDRALYSLLADGLVTQTADGRFALA
MLBr_01920|M.leprae_Br4923 DVLRAKDSPVTGTELNVTWLTNSIQLDRALDSLLADGLVTRTTDGRFALT
MSMEG_6083|M.smegmatis_MC2_155 DVLRDSTTPVTRAQLDVVWLSDPAQRDRALDSLLVDGLVEQTADGRFALA
TH_0591|M.thermoresistible__bu DVLRNSSAPVERAALDMVWLTDTAQRDRALHSLLADGLVEQTPDGRFALA
MAB_0558c|M.abscessus_ATCC_199 DVLRGSSIPVTRAQLDVAWLSDITQRDRALDSLLVDGLVEQTADGRFALA
**** ** : *::.* :: * **** ***.*.** :* ** ***
Mflv_1438|M.gilvum_PYR-GCK GEGDQT-----------------------
Mvan_5346|M.vanbaalenii_PYR-1 GEAGS------------------------
Mb3620|M.bovis_AF2122/97 GEGF-------------------------
Rv3589|M.tuberculosis_H37Rv GEGF-------------------------
MMAR_5089|M.marinum_M GEA--------------------------
MUL_4165|M.ulcerans_Agy99 GEE--------------------------
MAV_0563|M.avium_104 GEE--------------------------
MLBr_01920|M.leprae_Br4923 GEG--------------------------
MSMEG_6083|M.smegmatis_MC2_155 GEGETGRPA--------------------
TH_0591|M.thermoresistible__bu GEGEPDQSVTSAPDSVDPDVVEPQANRRR
MAB_0558c|M.abscessus_ATCC_199 GEGG-------------------------
**