For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEVAESPTHPAYGRLRAVTDTASVLLADNPGLLTLEGTNTWVLRGPGSDELVIVDPGPDDDEHIARIAA LGRIALVLISHRHGDHTDGIDKLVERTGATVRSAGSGFLRGLGGELTDGEVIDAAGLRIKVMATPGHTAD SLCFLLDDAVLTADTVLGRGTTVLDKEDGSLTDYLESLRRLRGLGRRAVLPGHGPDLTDLESVAQGYLEH RRERLDQVRSAVRELGEDVSARRIVEHVYVDVDEKLWDAAEWSVQVQLNHLRSEPST
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
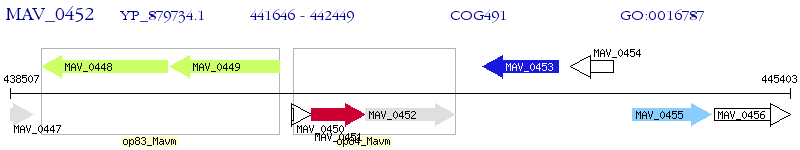
| M. avium 104 | MAV_0452 | - | - | 100% (267) | metallo-beta-lactamase family protein |
| M. avium 104 | MAV_4532 | - | 1e-10 | 29.17% (192) | metallo-beta-lactamase family protein |
| M. avium 104 | MAV_3461 | - | 3e-09 | 31.38% (188) | metallo-beta-lactamase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3701c | - | 1e-118 | 78.33% (263) | putative hydrolase |
| M. gilvum PYR-GCK | Mflv_1370 | - | 1e-114 | 78.35% (254) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv3677c | - | 1e-118 | 78.33% (263) | hydrolase |
| M. leprae Br4923 | MLBr_02303 | - | 1e-113 | 77.48% (262) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0414 | - | 4e-97 | 65.25% (259) | beta-lactamase-like hydrolase |
| M. marinum M | MMAR_5165 | - | 1e-121 | 81.20% (266) | hypothetical protein MMAR_5165 |
| M. smegmatis MC2 155 | MSMEG_6190 | - | 1e-113 | 77.52% (258) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_2128 | - | 1e-114 | 78.26% (253) | POSSIBLE HYDROLASE |
| M. ulcerans Agy99 | MUL_4251 | - | 1e-121 | 81.20% (266) | hypothetical protein MUL_4251 |
| M. vanbaalenii PYR-1 | Mvan_5436 | - | 1e-113 | 77.56% (254) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1370|M.gilvum_PYR-GCK -------MEHPAYNVLRPVTDTASVLLCENPGLMTLDGTNTWVLRGPGSD
Mvan_5436|M.vanbaalenii_PYR-1 -------MEHPAYGLLRPVTDTASVLLCENPGLMTLDGTNTWVLRGPGSD
MSMEG_6190|M.smegmatis_MC2_155 ----MSDLQHPAYGLLRPVTGSASVLLCNNPGLMTLEGTNTWVLRAPRSD
TH_2128|M.thermoresistible__bu ----VSDLEHPAYGLLRPVTETASVLLADNPGLMTLDGTNTWVLRGPGSD
MMAR_5165|M.marinum_M MSETAGSLTHPAYGRLRPVTDLASVLVADNPGLLTLEGTNTWVLRGPRSD
MUL_4251|M.ulcerans_Agy99 MSETAGSLTHPAYGRLRPVTDLASVLVADNPGLLTLEGTNTWVLRGPRSD
MAV_0452|M.avium_104 MTEVAESPTHPAYGRLRAVTDTASVLLADNPGLLTLEGTNTWVLRGPGSD
Mb3701c|M.bovis_AF2122/97 MSKTAESLTHPAYGQLRAVTDTASVLLADNPGLLTLDGTNTWVLRGPLSD
Rv3677c|M.tuberculosis_H37Rv MSKTAESLTHPAYGQLRAVTDTASVLLADNPGLLTLDGTNTWVLRGPLSD
MLBr_02303|M.leprae_Br4923 MAEVVKWLAHPAYGRLRAVTDTASVLLADNPGLLTLEGTNTWVLRGPGSG
MAB_0414|M.abscessus_ATCC_1997 --------MHPAYGQLRPVTETASVLLANNPGMMTLEGTNTWVLRAPGSD
****. **.** ****:.:***::**:********.* *.
Mflv_1370|M.gilvum_PYR-GCK EMVVVDPGP----DDKDEHIERLAALGRISLVLISHRHGDHTGGIDRLVD
Mvan_5436|M.vanbaalenii_PYR-1 EMVVVDPGP----DDRDEHIERLAALGPIPLVLISHRHADHTGGIDRIVD
MSMEG_6190|M.smegmatis_MC2_155 EIVVVDPGP----DD-DEHIARVAELGTISLVLISHKHEDHTGGIDKIVE
TH_2128|M.thermoresistible__bu EMVVVDPGP----DD-ADHIARLAELGPIPLVLISHKHEDHTGGIDKIVD
MMAR_5165|M.marinum_M ELVVVDPGP----AD-DEHIARIAALGRIALVLISHRHGDHTDGIDKLFE
MUL_4251|M.ulcerans_Agy99 ELVVVDPGP----AD-DEHIARIAALGRIALVLISHRHGDHTDGIDKLFE
MAV_0452|M.avium_104 ELVIVDPGP----DD-DEHIARIAALGRIALVLISHRHGDHTDGIDKLVE
Mb3701c|M.bovis_AF2122/97 ELVVVDPGP----DD-DEHLARVAALGRIALVLISHRHGDHTSGIDKLVA
Rv3677c|M.tuberculosis_H37Rv ELVVVDPGP----DD-DEHLARVAALGRIALVLISHRHGDHTSGIDKLVA
MLBr_02303|M.leprae_Br4923 ELVIVDPGP----GD--EHIVQVATLGRIALVLISHRHPDHTDGIDKLVE
MAB_0414|M.abscessus_ATCC_1997 EIVIVDPGPGVATGDPDVHVEQLAKIGKVALVLVSHRHFDHTGGVDRLVE
*:*:***** * *: ::* :* :.***:**:* ***.*:*::.
Mflv_1370|M.gilvum_PYR-GCK MTGAVVRSVGSGFQRGLGGTLTDGEVIDAAGLAITVMATPGHTADSVSFL
Mvan_5436|M.vanbaalenii_PYR-1 LTGAVVRSVGSGFQRGLGGTLTDGEVIDAAGLKITVMATPGHTADSMSFL
MSMEG_6190|M.smegmatis_MC2_155 RTGAVVRSVGSGFLRGLGGPLTDGEVIDAAGLRITVMATPGHTADSLSFV
TH_2128|M.thermoresistible__bu ATGAVVRSVGSGFLRGLGGPLTDGEVIEAAGLRITVLATPGHTVDSLSFV
MMAR_5165|M.marinum_M RTGAPVRAADPQFLRGDPVALSDGEVIDAGGLKITVLATPGHTADSLSFV
MUL_4251|M.ulcerans_Agy99 RTGAPVRAADPQFLRGDPVALSDGEVIDAGGLKITVLATPGHTADSLSFV
MAV_0452|M.avium_104 RTGATVRSAGSGFLRGLGGELTDGEVIDAAGLRIKVMATPGHTADSLCFL
Mb3701c|M.bovis_AF2122/97 LTGAPVRAADPQFLRRDGETLTDGEVIDVAGLTITVLATPGHTADSLSFV
Rv3677c|M.tuberculosis_H37Rv LTGAPVRAADPQFLRRDGETLTDGEVIDVAGLTITVLATPGHTADSLSFV
MLBr_02303|M.leprae_Br4923 LTGAPVCAADPQFLRGDGVTLTDREVIDAGGLTITVLATPGHTADSMSFV
MAB_0414|M.abscessus_ATCC_1997 LTGAPARAVDPAWLRGDSVALTDGERIDVAGLSISVLATPGHSDDSVSFV
*** . :... : * *:* * *:..** *.*:*****: **:.*:
Mflv_1370|M.gilvum_PYR-GCK VDD-----AVLTADTVLGRGTTVIDSEDGDLGDYLESLRRLRGLGHLTVL
Mvan_5436|M.vanbaalenii_PYR-1 VGD-----AVLTADTILGRGTTVIDSEDGDLGDYLESLRRLHGLGHRRVL
MSMEG_6190|M.smegmatis_MC2_155 LDDGDGPGAVLTADTVLGRGTTVIDTEDGSLRDYLDSLHRLKGLGERVVL
TH_2128|M.thermoresistible__bu LDD-----AVLTADTVLGRGTTVIDDEDGSLAAYLESLRRLRGLGRRVVL
MMAR_5165|M.marinum_M LED-----AVLTADSVLGRGTTVLDKQDGSLSEYLESLHRLRGLGPRMVL
MUL_4251|M.ulcerans_Agy99 LED-----AVLTADSVLGRGTTVLDKQDGSLSEYLESLHRLRGLGPRMVL
MAV_0452|M.avium_104 LDD-----AVLTADTVLGRGTTVLDKEDGSLTDYLESLRRLRGLGRRAVL
Mb3701c|M.bovis_AF2122/97 LDD-----AVLTADTVLGCGTTVIDKEDGSLADYLESLHRLRGLGRRTVL
Rv3677c|M.tuberculosis_H37Rv LDD-----AVLTADTVLGCGTTVIDKEDGSLADYLESLHRLRGLGRRTVL
MLBr_02303|M.leprae_Br4923 LAD-----AVLTADSVLGHGTTVIDPDDGSLADYLESLHRLSGLGRRTVL
MAB_0414|M.abscessus_ATCC_1997 LDD-----AVLTADTILGRGTTVIAQDGGGLGDYLESLKRLEGLGKRTVL
: * ******::** ****: :.*.* **:**:** *** **
Mflv_1370|M.gilvum_PYR-GCK PGHGPELDDMAAVSEFYLAHREERLAQVRGALGALGEDATARQVVEHVYT
Mvan_5436|M.vanbaalenii_PYR-1 PGHGPELDDVVVVSQDYLTHREERLEQVRAALRELGEDATARQVVEHVYT
MSMEG_6190|M.smegmatis_MC2_155 PGHGPELGDLSGVAQMYLAHREERLEQVREALRELGDDATARQVVEHVYT
TH_2128|M.thermoresistible__bu PGHGPELPDLAAVSDMYLAHREERLEQVRAALRELGEDATARQVVEHVYT
MMAR_5165|M.marinum_M PGHGPDLADIRIVAQGYLAHRQERLEQVRSALRELGEQASARQIVEHVYV
MUL_4251|M.ulcerans_Agy99 PGHGPDLADIRIVAQGYLAHRQERLEQVRSALRELGEQASARQIVEHVYV
MAV_0452|M.avium_104 PGHGPDLTDLESVAQGYLEHRRERLDQVRSAVRELGEDVSARRIVEHVYV
Mb3701c|M.bovis_AF2122/97 PGHGPDLLDLEAIASGYLLHRHERLEQIRAALRDLGDDATVREVVEHVYL
Rv3677c|M.tuberculosis_H37Rv PGHGPDLLDLEAIASGYLLHRHERLEQIRAALRDLGDDATVREVVEHVYL
MLBr_02303|M.leprae_Br4923 PGHGPDLANLESVASGYLVHRQERLEQVRAALRELGDDATARQVVEHVYV
MAB_0414|M.abscessus_ATCC_1997 PGHGPELSDLSAASAEYLAHREQRLDQVRAALAALGEDASARQVVEFVYT
*****:* :: : ** **.:** *:* *: **::.:.*.:**.**
Mflv_1370|M.gilvum_PYR-GCK DVDEKLWDAAEWSVQAQLNYLRR------
Mvan_5436|M.vanbaalenii_PYR-1 DVDEKLWDAAESSVQAQLDHLRR------
MSMEG_6190|M.smegmatis_MC2_155 DVDEKLWDAAEKSVQAQLDYLRH------
TH_2128|M.thermoresistible__bu DVDEKLWDAAESSVRAQLDYLRFRRGHDR
MMAR_5165|M.marinum_M DVDEKLWDAAEWSVQVQLNHLHQAGSPS-
MUL_4251|M.ulcerans_Agy99 DVDEKLWDAAEWSVQVQLNHLHQAGSQS-
MAV_0452|M.avium_104 DVDEKLWDAAEWSVQVQLNHLRSEPST--
Mb3701c|M.bovis_AF2122/97 DVDEKLWNAAEWSVQAQLDYLRTR-----
Rv3677c|M.tuberculosis_H37Rv DVDEKLWNAAEWSVQAQLDYLRTR-----
MLBr_02303|M.leprae_Br4923 DVDEKFWDAAEWSVQAQLDYLRR------
MAB_0414|M.abscessus_ATCC_1997 DVDPSLWGAAEWSVQAQLDYLRTV-----
*** .:*.*** **:.**::*: