For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAEFAEFIAAIDQGTTSTRCMIFDHQGAEVARHQLEHEQILPRAGWVEHDPIEIWERTSSVLTSVLNRAN LSAENLAALGITNQRETTLVWNRKTGRPYYNAIVWQDTRTDRIASALDRDGRGQVIRRKAGLPPATYFSG AKLQWILDNVDGVREAAERGDALFGTADSWVLWQLTGGPRGGVHATDVTNASRTMLMDLETLDWDDELLS FFTIPRAMLPEIGPSSSPRPFGVTSDTGPAGGRIPITAVLGDQHAAMVGQVCLAEGEAKNTYGTGNFLLL NTGESIVRSEHGLLTTVCYQFGDAKPVYALEGSIAVTGAAVQWLRDQLGIISGAAQSESLARQVDDNGGV YFVPAFSGLFAPYWRSDARGAIVGLSRFDTNAHLARATLEAICYQSRDVVDAMAADSGVRLEVLKVDGGI TGNDLCMQIQADVLGVDVVRPVVAETTALGAAYAAGLAVGFWADPGELRANWREDKRWTPAWSDEQRAAG YAGWRKAVQRTLDWADVT
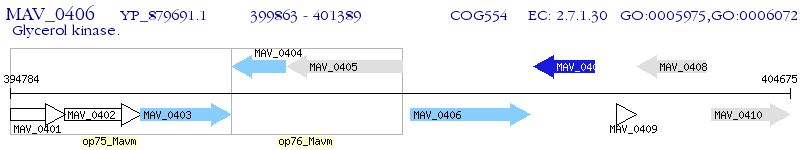
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0406 | glpK | - | 100% (508) | glycerol kinase |
| M. avium 104 | MAV_4437 | - | 2e-10 | 24.74% (481) | carbohydrate kinase, FGGY family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3722c | glpKa | 5e-92 | 86.26% (182) | glycerol kinase |
| M. gilvum PYR-GCK | Mflv_1327 | glpK | 0.0 | 85.09% (503) | glycerol kinase |
| M. tuberculosis H37Rv | Rv3696c | glpK | 0.0 | 87.60% (508) | glycerol kinase |
| M. leprae Br4923 | MLBr_02314 | glpK | 0.0 | 86.61% (508) | glycerol kinase |
| M. abscessus ATCC 19977 | MAB_0382 | glpK | 0.0 | 80.12% (503) | glycerol kinase |
| M. marinum M | MMAR_5208 | glpK | 0.0 | 87.60% (508) | glycerol kinase GlpK |
| M. smegmatis MC2 155 | MSMEG_6229 | glpK | 0.0 | 83.90% (503) | glycerol kinase |
| M. thermoresistible (build 8) | TH_1719 | glpK | 0.0 | 81.10% (508) | PROBABLE GLYCEROL KINASE GLPK (ATP:GLYCEROL |
| M. ulcerans Agy99 | MUL_4281 | glpK | 0.0 | 87.20% (508) | glycerol kinase |
| M. vanbaalenii PYR-1 | Mvan_5467 | glpK | 0.0 | 86.08% (503) | glycerol kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1327|M.gilvum_PYR-GCK ------------MADFVAAIDQGTTSTRCMIFDHDGAEVGRHQLEHEQIL
Mvan_5467|M.vanbaalenii_PYR-1 ------------MADFVAAIDQGTTSTRCMIFDHDGAEVGRHQLEHEQIL
MSMEG_6229|M.smegmatis_MC2_155 ------------MADFVAAIDQGTTSTRCMIFDHDGAEVGRHQLEHEQIL
TH_1719|M.thermoresistible__bu ------------VAEFVAAIDQGTTSTRCMIFDHAGTEVGRHQLEHQQIM
MAB_0382|M.abscessus_ATCC_1997 ------------MADFVASIDQGTTSTRAMIFNHRGEEVGRHQLEHQQLL
MAV_0406|M.avium_104 ---------MAEFAEFIAAIDQGTTSTRCMIFDHQGAEVARHQLEHEQIL
MMAR_5208|M.marinum_M ---------MADSPDFVAAIDQGTTSTRCMIFDHQGAEIARHQLEHEQIL
MUL_4281|M.ulcerans_Agy99 ---------MADSPDFVAAIDQGTTSTRCMIFDHQGAEIARHQLVHEQIL
MLBr_02314|M.leprae_Br4923 ---------MAEFAEFIAAIDQGTTSTRCVIFDHNGAEVARHQLEHEQIL
Rv3696c|M.tuberculosis_H37Rv MSDAILGEQLAESSDFIAAIDQGTTSTRCMIFDHHGAEVARHQLEHEQIL
Mb3722c|M.bovis_AF2122/97 MSDAILGEQLAESSDFIAAIDQGTTSTRCMIFDHHGAEVARHQLEHEQIL
.:*:*:*********.:**:* * *:.**** *:*::
Mflv_1327|M.gilvum_PYR-GCK PKAGWVEHNPVEIWERTGSVLATALNATKLATTDLAALGITNQRETSLVW
Mvan_5467|M.vanbaalenii_PYR-1 PRAGWVEHNPVEIWERTGSVLATALNKTKLSTSDLVALGITNQRETSLVW
MSMEG_6229|M.smegmatis_MC2_155 PRAGWVEHNPVEIWERTASVVMTALNRTNLQASDLAALGITNQRETSLVW
TH_1719|M.thermoresistible__bu PQAGWVEHNPIEIWERTSAVIMSALNETRLSATDLAALGVTNQRETTLVW
MAB_0382|M.abscessus_ATCC_1997 PRAGWVEHNPVEIWERTWSVLATSLNVTGLSAGDLAAVGVTNQRETTLVW
MAV_0406|M.avium_104 PRAGWVEHDPIEIWERTSSVLTSVLNRANLSAENLAALGITNQRETTLVW
MMAR_5208|M.marinum_M PRAGWVEHNPVEIWERTASVLMSVLNATDLSPKNIAALGITNQRETTLVW
MUL_4281|M.ulcerans_Agy99 PRAGWVEHNPVEIWERTASVLMSVLNATDLSPKNIAALGITNQRETTLVW
MLBr_02314|M.leprae_Br4923 PRSGWVEHNPVEIWERTASVLMSVLNATNLSAKDIAALGITNQRETTLVW
Rv3696c|M.tuberculosis_H37Rv PRAGWVEHNPVEIWERTASVLISVLNATNLSPKDIAALGITNQRETTLVW
Mb3722c|M.bovis_AF2122/97 PRAGWVEHNPVEIWERTASVLISVLNATNLSPKDIAALGITNQRETTLVW
*::*****:*:****** :*: : ** : * . ::.*:*:******:***
Mflv_1327|M.gilvum_PYR-GCK NRHTGRPYYNAIVWQDTRTDRIASALDRDGRGDVIRRKAGLPPATYFSGG
Mvan_5467|M.vanbaalenii_PYR-1 NRHTGRPYYNAIVWQDTRTDRIASALDRDGRGDVIRQKAGLPPATYFSGG
MSMEG_6229|M.smegmatis_MC2_155 NRHTGRPYYNAIVWQDTRTDSIAAALDRDGRGDVIRRKAGLPPATYFSGG
TH_1719|M.thermoresistible__bu NKRTGRPYYNAIVWQDTRTDRIASALERDGRGEVIRQKAGLPPATYFSGG
MAB_0382|M.abscessus_ATCC_1997 NRHTGRPYCNAIVWQDTRTDKIAAALDRDGRGDIIRRKAGLPPATYFSGG
MAV_0406|M.avium_104 NRKTGRPYYNAIVWQDTRTDRIASALDRDGRGQVIRRKAGLPPATYFSGA
MMAR_5208|M.marinum_M NKRTGRPYYNAIVWQDTRTDRIASALDRDGRGDVIRRKAGLPPATYFSGG
MUL_4281|M.ulcerans_Agy99 NKRTGRPYYNAIVWQDTRTDRIASALDRDGRGDVIRRKAGLPPATYFSGG
MLBr_02314|M.leprae_Br4923 NRNTGRPYCNAIVWQDTRTDRIAVALDRDGRGEVIRRKAGLPPATYFSGG
Rv3696c|M.tuberculosis_H37Rv NRHTGRPYYNAIVWQDTRTDRIASALDRDGRGNLIRRKAGLPPATYFSGG
Mb3722c|M.bovis_AF2122/97 NRHTGRPYYNAIVWQDTRTDRIASALDRDGRGNLIRRKAGLPPATYFSGG
*:.***** *********** ** **:*****::**:************.
Mflv_1327|M.gilvum_PYR-GCK KLQWLLENVDGLRADAANGDALFGTTDTWVLWNLTGGHRGGVHVTDVTNA
Mvan_5467|M.vanbaalenii_PYR-1 KLQWLLENVDGLRADAEKGDALFGTTDTWVLWNLTGGHRGGVHVTDVTNA
MSMEG_6229|M.smegmatis_MC2_155 KIQWILDNVPGVREDAEKGDAIFGTADSWLVWNLTGGTRGGVHVTDVTNA
TH_1719|M.thermoresistible__bu KLQWMLENVDGLRADAEKGEALFGTPDTWLVWNLTGGVRGGRHITDVTNA
MAB_0382|M.abscessus_ATCC_1997 KLQWILENVEGVRRDAENGDALFGTPDSWVIWNLTGGVRSGVHVTDVTNA
MAV_0406|M.avium_104 KLQWILDNVDGVREAAERGDALFGTADSWVLWQLTGGPRGGVHATDVTNA
MMAR_5208|M.marinum_M KLQWILENVDGVRAAAERGDALFGTPDTWVLWNLTGGPRGGVHVTDVTNA
MUL_4281|M.ulcerans_Agy99 KLQWILEDVDGVRAAAERGDALFGTPDTWVLWNLTGGPRGGVHVTDVTNA
MLBr_02314|M.leprae_Br4923 KLQWILENVDGVGAAAENGEALFGTPDTWLLWNLTGGPRGGVHVTDVTNA
Rv3696c|M.tuberculosis_H37Rv KLQWILENVDGVRAAAENGDALFGTPDTWVLWNLTGGPRGGVHVTDVTNA
Mb3722c|M.bovis_AF2122/97 KLQWILENVDGVRAAAENGDALFGTPDTWVLWNLTGGPRGG---------
*:**:*::* *: * .*:*:***.*:*::*:**** *.*
Mflv_1327|M.gilvum_PYR-GCK SRTMLMNLETLDWDDELLGFFDIPRQMLPEIRPSSSPQPYGVTVETGPAD
Mvan_5467|M.vanbaalenii_PYR-1 SRTMLMNLETLDWDDELLGFFDIPRQMLPEIRPSSSPEPHGVTVDWGPAD
MSMEG_6229|M.smegmatis_MC2_155 SRTMLMNLETLDWDDELLSFFGIPRRMLPEIKPSSYPESYGITRDDGPLA
TH_1719|M.thermoresistible__bu SRTMLMNLETLDWDDELLSFFGIPRAMLPEIKPSSYPQSYGITLDDGPVA
MAB_0382|M.abscessus_ATCC_1997 SRTMLMNLETLDWDDELLSFFSIPRQMLPPIKASSPVEPFGFTTQLGPLG
MAV_0406|M.avium_104 SRTMLMDLETLDWDDELLSFFTIPRAMLPEIGPSSSPRPFGVTSDTGPAG
MMAR_5208|M.marinum_M SRTMLMNLETLDWDDELLSFFSIPRAMLPAIEPSSPLQPYGITQDTGPVG
MUL_4281|M.ulcerans_Agy99 SRTMLMNLETLDWDDELLSFFSIPRAMLPAIEPSSPLQPYGITQDTGPVG
MLBr_02314|M.leprae_Br4923 SRTMLMDLEKLDWDDELLSFFSIPRAMLPAIASSSPLQPYGVTLADGPVG
Rv3696c|M.tuberculosis_H37Rv SRTMLMDLETLDWDDELLSLFSIPRAMLPEIASSAPSEPYGVTLATGPVG
Mb3722c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_1327|M.gilvum_PYR-GCK GEIPITGILGDQQAAMVGQVCLDVGEAKNTYGTGNFLLLNTGEKIVRSDN
Mvan_5467|M.vanbaalenii_PYR-1 GEIPVTGILGDQQAAMVGQVCLDAGEAKNTYGTGNFLLLNTGENIVRSKN
MSMEG_6229|M.smegmatis_MC2_155 GQVPLTGILGDQQAAMVGQVCLEAGEAKNTYGTGNFLLLNTGEKIVRSDN
TH_1719|M.thermoresistible__bu GQVPLTGILGDQQAAMVGQVCLSAGEAKNTYGTGNFLLLNTGENIVRSKN
MAB_0382|M.abscessus_ATCC_1997 GEVPIAGDLGDQQAAMVGQVCLNPGEAKNTYGTGNFLLLNTGEELVHSSN
MAV_0406|M.avium_104 GRIPITAVLGDQHAAMVGQVCLAEGEAKNTYGTGNFLLLNTGESIVRSEH
MMAR_5208|M.marinum_M GEVPITGMLGDQHAAMVGQVCLDAGEAKNTYGTGNFLLLNTGETIVRSSN
MUL_4281|M.ulcerans_Agy99 GEVPITGMLGDQHAAMVGQVCLDAGEAKNTYGTGNFLLLNTGETIVRSSN
MLBr_02314|M.leprae_Br4923 GEVPITGVLGDQHAAMVGQVCLDAGEAKNTYGTGNFLLLNTGEAIVRSGN
Rv3696c|M.tuberculosis_H37Rv GEVPITGVLGDQHAAMVGQVCLAPGEAKNTYGTGNFLLLNTGETIVRSNN
Mb3722c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_1327|M.gilvum_PYR-GCK GLLTTVCYQFGDS-----KPVYALEGSIAVTGSAVQWLRDQLGIISGASQ
Mvan_5467|M.vanbaalenii_PYR-1 GLLTTVCYQFGDA-----KPVYALEGSIAVTGSAVQWLRDQLGIISGAAQ
MSMEG_6229|M.smegmatis_MC2_155 GLLTTVCYQFGDS-----KPVYALEGSIAVTGSAVQWLRDQLGIISGASQ
TH_1719|M.thermoresistible__bu GLITTVCYQFTGADGRPAEPVYALEGSIAVTGAAVQWLRDQLGIISGAAQ
MAB_0382|M.abscessus_ATCC_1997 GLLTTVCYQFGDN-----KPVYALEGSIAVTGSAVQWLRDQLGIISGASQ
MAV_0406|M.avium_104 GLLTTVCYQFGDA-----KPVYALEGSIAVTGAAVQWLRDQLGIISGAAQ
MMAR_5208|M.marinum_M GLLTTLCYQFGDA-----KPVYALEGSIAVTGSAVQWLRDQLGIISGASQ
MUL_4281|M.ulcerans_Agy99 GLLTTLCYQFGDA-----KPVYALEGSIAVTGSAVQWLRDQLGIISGASQ
MLBr_02314|M.leprae_Br4923 GLLTTVCYQFGDA-----KPVYALEGSIAVTGSAVQWLRDQLGIISGAAQ
Rv3696c|M.tuberculosis_H37Rv GLLTTVCYQFGNA-----KPVYALEGSIAVTGSAVQWLRDQLGIISGAAQ
Mb3722c|M.bovis_AF2122/97 ----CACHRCNQR---------------------------QPDHVDGS--
*:: * . :.*:
Mflv_1327|M.gilvum_PYR-GCK SESLARQVDDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHVAR
Mvan_5467|M.vanbaalenii_PYR-1 SESLARQVEDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHVAR
MSMEG_6229|M.smegmatis_MC2_155 SESLARQVSDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHLAR
TH_1719|M.thermoresistible__bu SESLARQVPDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHIAR
MAB_0382|M.abscessus_ATCC_1997 SEDLARQVEDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHLAR
MAV_0406|M.avium_104 SESLARQVDDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFDTNAHLAR
MMAR_5208|M.marinum_M SEALARQVSDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNNNAHLAR
MUL_4281|M.ulcerans_Agy99 SEALARQVSDNGGVYFVPAFSGLFAPYWRSDARGAIVGLSRFNNNAHLAR
MLBr_02314|M.leprae_Br4923 SESLARQVVDNGGVYFVPAFSGLFSPYWRSDARGAIVGLSRFNTNAHLAR
Rv3696c|M.tuberculosis_H37Rv SEALARQVPDNGGMYFVPAFSGLFAPYWRSDARGAIVGLSRFNTNAHLAR
Mb3722c|M.bovis_AF2122/97 ----------------------------RDAGLGRRAVVVVFDTSGHAAR
*. . * . : *:...* **
Mflv_1327|M.gilvum_PYR-GCK ATLEAICYQSRDVVDAMAADSGVPLEVLKVDGGITANDLCMQIQADVLGV
Mvan_5467|M.vanbaalenii_PYR-1 ATLEAICYQSRDVVDAMAADSGVHLEVLKVDGGITANDLCMQIQADVLGV
MSMEG_6229|M.smegmatis_MC2_155 ATLESICYQSRDVVDAMEADSGVHLEVLKVDGGITANALCMQIQADVLGV
TH_1719|M.thermoresistible__bu ATLEAICYQSRDVVDAMVADSGVRMESLKVDGGITANELCMQIQADVLGV
MAB_0382|M.abscessus_ATCC_1997 ATLEAICYQSREVVEAMEADSGVRMEVLKVDGGITANKLCMQIQADTLGV
MAV_0406|M.avium_104 ATLEAICYQSRDVVDAMAADSGVRLEVLKVDGGITGNDLCMQIQADVLGV
MMAR_5208|M.marinum_M ATLEAICYQSRDVVDAMEADSGVRLEVLKVDGGITSNDLCMQIQADVLGV
MUL_4281|M.ulcerans_Agy99 ATLEAICYQSRDVVDAMEADSGVRLEVLKVDGGITSNDLCMQIQADVLGV
MLBr_02314|M.leprae_Br4923 ATLEAICYQSRDVVDAMAADSGVRLDVLKVDGGITSNDLCMQIQADVLGV
Rv3696c|M.tuberculosis_H37Rv ATLEAICYQSRDVVDAMEADSGVRLQVLKVDGGITGNDLCMQIQADVLGV
Mb3722c|M.bovis_AF2122/97 DRIVG-------AVGALRCHAG--------------DRACRR--------
: . .* *: ..:* : * :
Mflv_1327|M.gilvum_PYR-GCK DVVKPVVAETTALGAAYAAGLAVGFWEGADDLRANWQEGRRWSPQWSDEQ
Mvan_5467|M.vanbaalenii_PYR-1 DVVRPVVAETTALGAAYAAGLAVGFWDGADDLRANWQEDKRWSPTWSDDQ
MSMEG_6229|M.smegmatis_MC2_155 DVVKPLVAETTALGAAYAAGLAVGFWENADDLRANWQEDKRWSPQWSDEQ
TH_1719|M.thermoresistible__bu DVIKPVVSETTALGAAYAAGLAVGFWEDADDLRTNWQEGRRWTPQWTDEQ
MAB_0382|M.abscessus_ATCC_1997 DVVKPVVAETTALGAAYAAGLAVGFWKDADDLRRNWQEDERWSSSITDEK
MAV_0406|M.avium_104 DVVRPVVAETTALGAAYAAGLAVGFWADPGELRANWREDKRWTPAWSDEQ
MMAR_5208|M.marinum_M DVVRPVVAETTALGAAYAAGLAVGFWADPSDLRANWQEDKRWTPQWTDDQ
MUL_4281|M.ulcerans_Agy99 DVVRPVVAETTALGAAYAAGLAVGFWADPSDLRANWQEDKRWTPQWTDDQ
MLBr_02314|M.leprae_Br4923 DVVRSVVAETTALGSAYAAGLAVGFWAGPSDLRANWQEDKRWTPTWGEDQ
Rv3696c|M.tuberculosis_H37Rv DVVRPVVAETTALGVAYAAGLAVGFWAAPSDLRANWREDKRWTPTWDDDE
Mb3722c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_1327|M.gilvum_PYR-GCK RAEGYAGWQKAVHRTLDWVDVE
Mvan_5467|M.vanbaalenii_PYR-1 RAAGYAGWRKAVQRTMDWVDVD
MSMEG_6229|M.smegmatis_MC2_155 REKGYAGWQKAVQRTLDWVEIE
TH_1719|M.thermoresistible__bu REAGYRGWKKAVQRTLDWVDVG
MAB_0382|M.abscessus_ATCC_1997 RAEGFAGWKKAVQRTLDGVDV-
MAV_0406|M.avium_104 RAAGYAGWRKAVQRTLDWADVT
MMAR_5208|M.marinum_M RAAGYAGWRKAVQRTLDWVDVS
MUL_4281|M.ulcerans_Agy99 RAAGYAGWRKAVQRTLDWVDVS
MLBr_02314|M.leprae_Br4923 RAAAYAGWRKAVQRTLDWVDVS
Rv3696c|M.tuberculosis_H37Rv RAAGYAGWRKAVQRTLDWVDVS
Mb3722c|M.bovis_AF2122/97 ----------------------