For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPAPIVADLRAESDALDALVAPLPPGRWADPTPAAGWTIAHQIGHLLWTDRVALIAVTDEPGFAEVLAG AAADPAGFVDKGAEELAAVAPAELLADWRATRDRLHRALLGVPAGRKLPWFGPPMSAASMATARLMETWA HGLDVADALGVTRPATDRLRSIAHLGVRTRDYAFAVNNLTPPAEPFLVELRGPRGDTWSWGPPEAAQRVT GSAEDFCFLVTQRRPLSALDITADGADAQRWLEIAQAFAGPPGPGR*
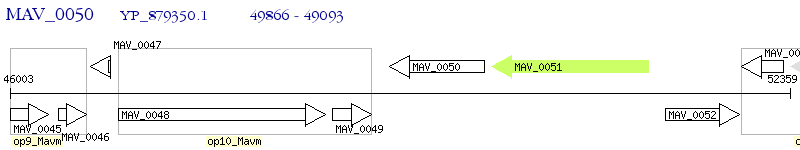
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0050 | - | - | 100% (257) | hypothetical protein MAV_0050 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0037c | - | 1e-124 | 83.66% (257) | hypothetical protein Mb0037c |
| M. gilvum PYR-GCK | Mflv_0848 | - | 1e-104 | 72.37% (257) | wyosine base formation |
| M. tuberculosis H37Rv | Rv0036c | - | 1e-124 | 83.66% (257) | hypothetical protein Rv0036c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4930 | - | 1e-100 | 69.72% (251) | hypothetical protein MAB_4930 |
| M. marinum M | MMAR_0051 | - | 1e-123 | 82.10% (257) | hypothetical protein MMAR_0051 |
| M. smegmatis MC2 155 | MSMEG_6923 | - | 1e-111 | 76.19% (252) | hypothetical protein MSMEG_6923 |
| M. thermoresistible (build 8) | TH_2156 | - | 3e-06 | 24.53% (159) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0050 | - | 1e-122 | 81.71% (257) | hypothetical protein MUL_0050 |
| M. vanbaalenii PYR-1 | Mvan_6059 | - | 1e-108 | 73.15% (257) | hypothetical protein Mvan_6059 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0848|M.gilvum_PYR-GCK -MSRMAAAAPLIAELRAESDELDALVADLTPQQWTLSTPAPGWTIAHQIA
Mvan_6059|M.vanbaalenii_PYR-1 ----MADAARVIDELRAESDDLDALVAALPAARWTLPTPAPGWTIAHQIG
Mb0037c|M.bovis_AF2122/97 ----MADPGPFVADLRAESDDLDALVAHLPADRWADPTPAPGWTIAHQIG
Rv0036c|M.tuberculosis_H37Rv ----MADPGPFVADLRAESDDLDALVAHLPADRWADPTPAPGWTIAHQIG
MMAR_0051|M.marinum_M ----MAGPGPIVADLEAESDDLDALVAPLPPQRWSELTPAPGWSIAHQIA
MUL_0050|M.ulcerans_Agy99 ----MAGPGPIVADLEAENDDLDALVAPLAPQRWSELTPAPGWTIAHQIA
MAV_0050|M.avium_104 ----MTGPAPIVADLRAESDALDALVAPLPPGRWADPTPAAGWTIAHQIG
MSMEG_6923|M.smegmatis_MC2_155 -----MAAEPIVADLRAESDALDALVADLPEAAWATDTPAQGWTIAHQIG
MAB_4930|M.abscessus_ATCC_1997 ---MTSTADAVIDDLRAESDELDALVAALPEQGWARDTPAAGWTIAHQIA
TH_2156|M.thermoresistible__bu VSITALDKTEILDGLFAVWDDITALGGRLTDAQWQTPTELPGWSVHDVLA
.: * * * : ** . *. * * **:: . :.
Mflv_0848|M.gilvum_PYR-GCK HLLWTDRVAVLAVTDEARFADVVAASAHDS-AFVDT-AAAELAVLPPAEL
Mvan_6059|M.vanbaalenii_PYR-1 HLMWTDRVALLSVTDEAGFAAVLAQASENPGGFVDA-AAEELALTSPGEL
Mb0037c|M.bovis_AF2122/97 HLLWTDRVALTAVTDEAGFAELMTAAAANPAGFVDD-AATELAAVSPAEL
Rv0036c|M.tuberculosis_H37Rv HLLWTDRVALTAVTDEAGFAELMTAAAANPAGFVDD-AATELAAVSPAEL
MMAR_0051|M.marinum_M HLLWTDRVALTAVTDEAGFADVLTAAAANPAGFVDE-GAEELAALPPAEL
MUL_0050|M.ulcerans_Agy99 HLLWTDRVALTAVTDEAGFADVLTAAAANPAGFVDE-GAEELAAMPPAEL
MAV_0050|M.avium_104 HLLWTDRVALIAVTDEPGFAEVLAGAAADPAGFVDK-GAEELAAVAPAEL
MSMEG_6923|M.smegmatis_MC2_155 HLLWTDQMALLAVTDEAGFAEAVARAATHPTGFVDE-GAAALAALPASQL
MAB_4930|M.abscessus_ATCC_1997 HLHWTDAQSLLAVTDPAVFAEELPKAMADPLGFVDK-AAEETAQIPPADL
TH_2156|M.thermoresistible__bu HLIGIESMLQGVETPDADIDVTTLEHVHNDIGALNERWIRALRGLEPAEL
** : * . : . . :: ..:*
Mflv_0848|M.gilvum_PYR-GCK LADWRTTRAALHDALLAVTDGRKLPWFGPPMSAASMA---TARLMETWAH
Mvan_6059|M.vanbaalenii_PYR-1 LADWRATRAALHDALSTVADGRKLPWFGPPMSAASMA---TARLMETWAH
Mb0037c|M.bovis_AF2122/97 LTDWRVTRGRLHEELLAVPDGRKLAWFGPPMSAASMA---TARLMETWAH
Rv0036c|M.tuberculosis_H37Rv LTDWRVTRGRLHEELLAVPDGRKLAWFGPPMSAASMA---TARLMETWAH
MMAR_0051|M.marinum_M LSDWRLTRGRLHEALLNVADGRKLPWFGPPMSAASMA---TARLMETWAH
MUL_0050|M.ulcerans_Agy99 LSDWRLTRGRLHEALLNVADGRKLPWFGPPMSAASMA---TARLMETWAH
MAV_0050|M.avium_104 LADWRATRDRLHRALLGVPAGRKLPWFGPPMSAASMA---TARLMETWAH
MSMEG_6923|M.smegmatis_MC2_155 LAEWRTTRNRLHDALLGVADGRKLPWFGPPMAAGSMA---TARLMETWAH
MAB_4930|M.abscessus_ATCC_1997 LIQWRSTRNQLHAALRAVPDGVKIPWYGPPMSAASMG---TARLMETWAH
TH_2156|M.thermoresistible__bu LDRWRSVTDERRAALTAMNDEDWSAVTATPAGPDSYGRFMRIRIFDCWMH
* ** . : * : . ..* .. * . *::: * *
Mflv_0848|M.gilvum_PYR-GCK GLDVADTLGTVRIPTSRLRSIAHIGVRTRDFAF----AVHGLTPPPEPFH
Mvan_6059|M.vanbaalenii_PYR-1 GLDVADALGVTRAPTARLRSIAHIGVRTRDFAF----TVHGLTPPPEPFH
Mb0037c|M.bovis_AF2122/97 GLDVADALGVIRPATQRLRSIAHLGVRTRDYAF----IVNNLTPPAEPFL
Rv0036c|M.tuberculosis_H37Rv GLDVADALGVIRPATQRLRSIAHLGVRTRDYAF----IVNNLTPPAEPFL
MMAR_0051|M.marinum_M GLDVADALGVTRPATDRLRSIAHLGVRTRDYAY----FVNNLAPPTEQFL
MUL_0050|M.ulcerans_Agy99 GLDVADALGVTRPATDRLRSIAHLGVRTRDYAY----FVNNLAPPTEQFL
MAV_0050|M.avium_104 GLDVADALGVTRPATDRLRSIAHLGVRTRDYAF----AVNNLTPPAEPFL
MSMEG_6923|M.smegmatis_MC2_155 GLDVADALGVRRPATDRLRSVAHIGVRTRDFAF----SVHGLTPPAEPFR
MAB_4930|M.abscessus_ATCC_1997 GLDVADALGVPLAPTPRIKSIAHLGVRTRDFAF----SVHGLPAPSEPFR
TH_2156|M.thermoresistible__bu EHDIRAVTGLTLGDDALDTPPARLALDEMAAAMGRVVGKLGKAPDGSRVR
*: . * . *::.: * . .. . .
Mflv_0848|M.gilvum_PYR-GCK VKLRAPDGSTWSWGTEDADQRVEG-----SAEHFCMLVTQRRPPAALDVV
Mvan_6059|M.vanbaalenii_PYR-1 VKLSAPHGAQWTWGPEDAAQRVTG-----SAEHFCMLATQRRPRAALDVV
Mb0037c|M.bovis_AF2122/97 VELRGPSGDTWSWGPSDAAQRVTG-----SAEDFCFLVTQRRALSTLDVN
Rv0036c|M.tuberculosis_H37Rv VELRGPSGDTWSWGPSDAAQRVTG-----SAEDFCFLVTQRRALSTLDVN
MMAR_0051|M.marinum_M VELRGPGGDIWSWGPADAAQRVTG-----PAEDFCFLVTQRRPLSSLHLT
MUL_0050|M.ulcerans_Agy99 VELRGPGGDLWSWGPADAAQRVTG-----PAEDFCFLVTQRRPLSSLHLT
MAV_0050|M.avium_104 VELRGPRGDTWSWGPPEAAQRVTG-----SAEDFCFLVTQRRPLSALDIT
MSMEG_6923|M.smegmatis_MC2_155 VELTGPSGELWVWGPEDAAQRVTG-----VAEDFCMLVTQRRAPSQLDVH
MAB_4930|M.abscessus_ATCC_1997 VELTGPDADTWTWGPEEAAQRVTG-----SALDFCYLVTQRRPRADLDLR
TH_2156|M.thermoresistible__bu FELLGPLARTIDVAVQDGRGRVVPGFGATGDGDPGPTTVISLDAWVFTRL
.:* .* . . :. ** . .. :
Mflv_0848|M.gilvum_PYR-GCK ATGPDAQRWLTIAQAFAGPPGAGRD---------
Mvan_6059|M.vanbaalenii_PYR-1 AVGPDAEKWLTIAQAFAGPPGPGRG---------
Mb0037c|M.bovis_AF2122/97 AVGEDAQRWLTIAQAFAGPPGRGR----------
Rv0036c|M.tuberculosis_H37Rv AVGEDAQRWLTIAQAFAGPPGRGR----------
MMAR_0051|M.marinum_M ATGEDARQWLQIAQAFAGPPGEGR----------
MUL_0050|M.ulcerans_Agy99 ATGEDARQWLQIAQAFAGPPGEGR----------
MAV_0050|M.avium_104 ADGADAQRWLEIAQAFAGPPGPGR----------
MSMEG_6923|M.smegmatis_MC2_155 ATGPDAQTWLTIAQAFAGPPGAGRG---------
MAB_4930|M.abscessus_ATCC_1997 ATGQDAEQWLSIAQAFAGPPGAGRG---------
TH_2156|M.thermoresistible__bu AGGRSTPERHPGAIGYRGDEAVGRRIVENLNYVI
* * .: * .: * . **