For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DDSVRVQVLAEIFISRIAWFSEKIERAQIDTPRRSTGHRRVFELFNEYVDQVRARVNEDLAADDIEPTDL ARQLRLLMTQFVGLHRTFDDLFSRDRSRVPQSLPFFVQEACRQWELPAADTVITVGKPGNFTTRNRSSWN TMFLYGSPSNILSIAVPEHEGTQARWLPITLGHELAHHYLLAGNIPELEEIEAQIAEQFAAALKGEDGQG AESTFADQWRAEEVLHEWVEELVCDAYAVDLFGPAAVAASLEFLHSLGDPSAPTKTHPPTLFRAELMLAW LADVTAGAPDHGTLSLQYLGDEKITHPQSALLCDAIRTQSQAIWDAVRSWSKERSYCATEREPHVDALAR LLSQGIPGAFTPDTPHDVIDIVNAYWRARSAGTQFPIDRLALKAIDDLYFIKLWTSADGAIHEDHVSCDW VGGPGVLTNADFRTRINNGDDNQISITPMLPGALGPASVDLRLSNQFIVFEQTSNASFNALEMLQDPRSM QSRLEKDWGDEFYLHPGQMVLASTLEYIVLPSDLCAQVITRSSYGRLGLLSATAVQVHPGFRGCLTLELV NLGKMPLAITPGERVAQLMFFGVSSPHAPAAETARKYECPTGPEFSKIKTDPELNTLRNIRNYSTRTTTA VNVEATRGRPEKSQGETGGNTRG*
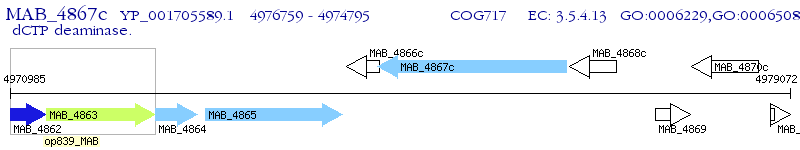
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4867c | - | - | 100% (654) | dCTP deaminase/DeoxyUTP pyrophosphatase |
| M. abscessus ATCC 19977 | MAB_4297c | dcd | 5e-24 | 34.95% (186) | deoxycytidine triphosphate deaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0329 | dcd | 3e-23 | 34.41% (186) | deoxycytidine triphosphate deaminase |
| M. gilvum PYR-GCK | Mflv_0291 | dcd | 1e-24 | 35.48% (186) | deoxycytidine triphosphate deaminase |
| M. tuberculosis H37Rv | Rv0321 | dcd | 4e-23 | 34.41% (186) | deoxycytidine triphosphate deaminase |
| M. leprae Br4923 | MLBr_02507 | dcd | 1e-23 | 36.02% (186) | deoxycytidine triphosphate deaminase |
| M. marinum M | MMAR_0600 | dcd | 2e-24 | 36.56% (186) | deoxycytidine triphosphate deaminase, Dcd |
| M. avium 104 | MAV_4828 | dcd | 1e-24 | 36.56% (186) | deoxycytidine triphosphate deaminase |
| M. smegmatis MC2 155 | MSMEG_0678 | dcd | 7e-23 | 33.87% (186) | deoxycytidine triphosphate deaminase |
| M. thermoresistible (build 8) | TH_1210 | dcd | 1e-23 | 35.48% (186) | PROBABLE DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE DCD (DCTP |
| M. ulcerans Agy99 | MUL_0560 | dcd | 7e-25 | 36.56% (186) | deoxycytidine triphosphate deaminase |
| M. vanbaalenii PYR-1 | Mvan_0589 | dcd | 2e-24 | 35.48% (186) | deoxycytidine triphosphate deaminase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 MDDSVRVQVLAEIFISRIAWFSEKIERAQIDTPRRSTGHRRVFELFNEYV
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 DQVRARVNEDLAADDIEPTDLARQLRLLMTQFVGLHRTFDDLFSRDRSRV
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 PQSLPFFVQEACRQWELPAADTVITVGKPGNFTTRNRSSWNTMFLYGSPS
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 NILSIAVPEHEGTQARWLPITLGHELAHHYLLAGNIPELEEIEAQIAEQF
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 AAALKGEDGQGAESTFADQWRAEEVLHEWVEELVCDAYAVDLFGPAAVAA
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 SLEFLHSLGDPSAPTKTHPPTLFRAELMLAWLADVTAGAPDHGTLSLQYL
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 GDEKITHPQSALLCDAIRTQSQAIWDAVRSWSKERSYCATEREPHVDALA
Mb0329|M.bovis_AF2122/97 --------------------------------------------------
Rv0321|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4828|M.avium_104 --------------------------------------------------
MLBr_02507|M.leprae_Br4923 --------------------------------------------------
Mflv_0291|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0589|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0678|M.smegmatis_MC2_155 --------------------------------------------------
TH_1210|M.thermoresistible__bu --------------------------------------------------
MMAR_0600|M.marinum_M --------------------------------------------------
MUL_0560|M.ulcerans_Agy99 --------------------------------------------------
MAB_4867c|M.abscessus_ATCC_199 RLLSQGIPGAFTPDTPHDVIDIVNAYWRARSAGTQFPIDRLALKAIDDLY
Mb0329|M.bovis_AF2122/97 -------------------------MLLSDRDLRAEISSG---RLGIDPF
Rv0321|M.tuberculosis_H37Rv -------------------------MLLSDRDLRAEISSG---RLGIDPF
MAV_4828|M.avium_104 -------------------------MLLSDRDLRAEITAG---RLGIEPF
MLBr_02507|M.leprae_Br4923 -------------------------MLLSDRDLRAEITAG---RFSIDPF
Mflv_0291|M.gilvum_PYR-GCK -------------------------MLLSDRDIRAEFQAG---RLGLDPF
Mvan_0589|M.vanbaalenii_PYR-1 -------------------------MLLSDRDIRAEITAG---RLGVDPF
MSMEG_0678|M.smegmatis_MC2_155 -------------------------MLLSDRDIRAEIAAK---RLALEPF
TH_1210|M.thermoresistible__bu LNYTGTAQQVLLVMKASTPSRYAHRVLLSDRDIRAEIAAG---RLALEPF
MMAR_0600|M.marinum_M -------------------------MLLSDRDLRAEISAG---RLGIDPF
MUL_0560|M.ulcerans_Agy99 -------------------------MLLSDRDLRAEISAG---RLGIDPF
MAB_4867c|M.abscessus_ATCC_199 FIKLWTSADGAIHEDHVSCDWVGGPGVLTNADFRTRINNGDDNQISITPM
:*:: *:*:.: ::.: *:
Mb0329|M.bovis_AF2122/97 DDTLVQPSSIDVRLDCLFRVFNNTR---YTHIDPAKQQDELTSLVQPVDG
Rv0321|M.tuberculosis_H37Rv DDTLVQPSSIDVRLDCLFRVFNNTR---YTHIDPAKQQDELTSLVQPVDG
MAV_4828|M.avium_104 DDALVQPSSIDVRLDCMFRVFNNTR---YTHIDPAKQQDELTSLVEPQDG
MLBr_02507|M.leprae_Br4923 DDTLVQPSSIDVRLDCMFRVFNNTR---YTHIDPARQQDELTSLVELVDG
Mflv_0291|M.gilvum_PYR-GCK DDSLIQPSSVDVRLDNLFRVFNNTR---YTHIDPAQRQDDLTSLVEPKEG
Mvan_0589|M.vanbaalenii_PYR-1 DDSLVQPSSVDVRLDNLFRVFNNTR---YTHIDPAQRQDDLTSLVEPKEG
MSMEG_0678|M.smegmatis_MC2_155 DDALVQPSSIDVRLDRMFRVFNNTR---YTHIDPAMQQDELTTLVEPAEG
TH_1210|M.thermoresistible__bu DDSLVQPSSIDVRLDNLFRVFNNTR---YTHIDPSIRQDELTSLVQAEDG
MMAR_0600|M.marinum_M DDSLVQPSSVDVRLDCMFRVFNNTR---YTHIDPAQRQDELTSVVEPTKG
MUL_0560|M.ulcerans_Agy99 DDSLVQPSSVDVRLDCMFRVFNNTR---YTHIDPAQRQDELTSAVEPTKG
MAB_4867c|M.abscessus_ATCC_199 LPGALGPASVDLRLSNQFIVFEQTSNASFNALEMLQDPRSMQSRLEKDWG
: *:*:*:**. * **::* :. :: .: : :: *
Mb0329|M.bovis_AF2122/97 EPFVLHPGEFVLGSTLELFTLPDNLAGRLEGKSSLGRLGLLTHSTAGFID
Rv0321|M.tuberculosis_H37Rv EPFVLHPGEFVLGSTLELFTLPDNLAGRLEGKSSLGRLGLLTHSTAGFID
MAV_4828|M.avium_104 EPFVLHPGEFVLGSTLELITLPDDLAGRLEGKSSLGRLGLLTHSTAGFID
MLBr_02507|M.leprae_Br4923 EPFVLHPGGFVLGSTLELFTLPEDLAGRLEGKSSLGRLGLLTHSTAGFID
Mflv_0291|M.gilvum_PYR-GCK EPFVLHPGEFVLGATLERCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFID
Mvan_0589|M.vanbaalenii_PYR-1 EPFVLHPGEFVLGATLERCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFID
MSMEG_0678|M.smegmatis_MC2_155 EPFVLHPGEFVLGSTLELCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFID
TH_1210|M.thermoresistible__bu EPFVLHPGEFVLGSTLELCSLPDDLAGRLEGKSSLGRLGLLTHSTAGFID
MMAR_0600|M.marinum_M DPFVLHPGEFVLGSTLEIFSLPGDLAGRLEGKSSLGRLGLLTHSTAGFID
MUL_0560|M.ulcerans_Agy99 DPFVLHPGEFVLGSTLEIFSLPGDLAGRLEGKSSLGRLGLLTHSTAGFID
MAB_4867c|M.abscessus_ATCC_199 DEFYLHPGQMVLASTLEYIVLPSDLCAQVITRSSYGRLGLLS-ATAVQVH
: * **** :**.:*** ** :*..:: :** ******: :** :.
Mb0329|M.bovis_AF2122/97 PGFSGHITLELSNVANLPITLWPGMKIGQLCMLRLTSPSE--------HP
Rv0321|M.tuberculosis_H37Rv PGFSGHITLELSNVANLPITLWPGMKIGQLCMLRLTSPSE--------HP
MAV_4828|M.avium_104 PGFSGHITLELSNVANLPITLWPGMKIGQLCILRLTSPAE--------HP
MLBr_02507|M.leprae_Br4923 PGFCGHITLELSNVANLPITLWPGMKIGQLCVLRLTSPAE--------HP
Mflv_0291|M.gilvum_PYR-GCK PGFSGHITLELSNVANLPITLWPGMKIGQLCLLRLTSPAQ--------HP
Mvan_0589|M.vanbaalenii_PYR-1 PGFSGHITLELSNVANLPITLWPGMKIGQLCLLRLTSPAE--------HP
MSMEG_0678|M.smegmatis_MC2_155 PGFSGHITLELSNVANLPITLWPGMKIGQLCLLRLTSPAE--------NP
TH_1210|M.thermoresistible__bu PGFSGHITLELSNVANLPITLWPGMKIGQLCLLRLTSPAE--------FP
MMAR_0600|M.marinum_M PGFSGHITLELSNVANLPITLWPGMKIGQLCILKLTSPSE--------HP
MUL_0560|M.ulcerans_Agy99 PGFSGHITLELSNVANLPITLWPGMKIGQLCILKLTSPSE--------HP
MAB_4867c|M.abscessus_ATCC_199 PGFRGCLTLELVNLGKMPLAITPGERVAQLMFFGVSSPHAPAAETARKYE
*** * :**** *:.::*::: ** ::.** .: ::**
Mb0329|M.bovis_AF2122/97 YGSSRAGSKYQGQRGPTPSRSCQNFIRST---------------------
Rv0321|M.tuberculosis_H37Rv YGSSRAGSKYQGQRGPTPSRSYQNFIRST---------------------
MAV_4828|M.avium_104 YGSTRVGSKYQGQRGPTPSRSYQNFITST---------------------
MLBr_02507|M.leprae_Br4923 YGSASAGSKYQGQRGPTPSRSYENFIKNT---------------------
Mflv_0291|M.gilvum_PYR-GCK YGSSQVGSKYQGQRGPTPSKSYQNFVKSN---------------------
Mvan_0589|M.vanbaalenii_PYR-1 YGSSSVGSKYQGQRGPTPSRSYQNFVKND---------------------
MSMEG_0678|M.smegmatis_MC2_155 YGSAAVGSKYQGQRGPTPSRSHLNFIKS----------------------
TH_1210|M.thermoresistible__bu YGSTQAGSKYQGQRGPTPSRSYQNFVKSIRPQP-----------------
MMAR_0600|M.marinum_M YGSSGVGSKYQGQRGPTPSRSYQNFIRST---------------------
MUL_0560|M.ulcerans_Agy99 YGSSGVGSKYQGQRGPTPSRSYQNFIRST---------------------
MAB_4867c|M.abscessus_ATCC_199 CPTGPEFSKIKTDPELNTLRNIRNYSTRTTTAVNVEATRGRPEKSQGETG
: ** : : .. :. *:
Mb0329|M.bovis_AF2122/97 -----
Rv0321|M.tuberculosis_H37Rv -----
MAV_4828|M.avium_104 -----
MLBr_02507|M.leprae_Br4923 -----
Mflv_0291|M.gilvum_PYR-GCK -----
Mvan_0589|M.vanbaalenii_PYR-1 -----
MSMEG_0678|M.smegmatis_MC2_155 -----
TH_1210|M.thermoresistible__bu -----
MMAR_0600|M.marinum_M -----
MUL_0560|M.ulcerans_Agy99 -----
MAB_4867c|M.abscessus_ATCC_199 GNTRG