For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLLSDRDLRAEISSGRLGIDPFDDTLVQPSSIDVRLDCLFRVFNNTRYTHIDPAKQQDELTSLVQPVDGE PFVLHPGEFVLGSTLELFTLPDNLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGHITLELSNVANLPITL WPGMKIGQLCMLRLTSPSEHPYGSSRAGSKYQGQRGPTPSRSYQNFIRST
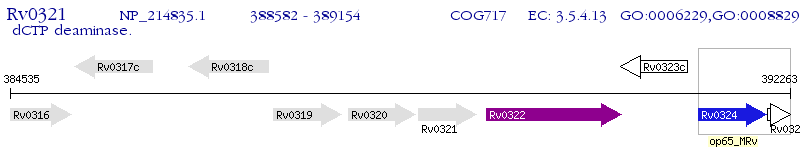
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0321 | dcd | - | 100% (190) | deoxycytidine triphosphate deaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0329 | dcd | 1e-107 | 99.47% (190) | deoxycytidine triphosphate deaminase |
| M. gilvum PYR-GCK | Mflv_0291 | dcd | 4e-96 | 85.71% (189) | deoxycytidine triphosphate deaminase |
| M. leprae Br4923 | MLBr_02507 | dcd | 1e-100 | 90.00% (190) | deoxycytidine triphosphate deaminase |
| M. abscessus ATCC 19977 | MAB_4297c | dcd | 2e-94 | 86.63% (187) | deoxycytidine triphosphate deaminase |
| M. marinum M | MMAR_0600 | dcd | 1e-100 | 90.00% (190) | deoxycytidine triphosphate deaminase, Dcd |
| M. avium 104 | MAV_4828 | dcd | 1e-102 | 92.63% (190) | deoxycytidine triphosphate deaminase |
| M. smegmatis MC2 155 | MSMEG_0678 | dcd | 6e-94 | 86.24% (189) | deoxycytidine triphosphate deaminase |
| M. thermoresistible (build 8) | TH_1210 | dcd | 1e-95 | 87.83% (189) | PROBABLE DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE DCD (DCTP |
| M. ulcerans Agy99 | MUL_0560 | dcd | 1e-100 | 90.00% (190) | deoxycytidine triphosphate deaminase |
| M. vanbaalenii PYR-1 | Mvan_0589 | dcd | 5e-97 | 87.30% (189) | deoxycytidine triphosphate deaminase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0678|M.smegmatis_MC2_155 -------------------------MLLSDRDIRAEIAAKRLALEPFDDA
TH_1210|M.thermoresistible__bu LNYTGTAQQVLLVMKASTPSRYAHRVLLSDRDIRAEIAAGRLALEPFDDS
MAB_4297c|M.abscessus_ATCC_199 -------------------------MLLSDRDIRAELDAGRLGIEPLDAA
Mflv_0291|M.gilvum_PYR-GCK -------------------------MLLSDRDIRAEFQAGRLGLDPFDDS
Mvan_0589|M.vanbaalenii_PYR-1 -------------------------MLLSDRDIRAEITAGRLGVDPFDDS
Rv0321|M.tuberculosis_H37Rv -------------------------MLLSDRDLRAEISSGRLGIDPFDDT
Mb0329|M.bovis_AF2122/97 -------------------------MLLSDRDLRAEISSGRLGIDPFDDT
MLBr_02507|M.leprae_Br4923 -------------------------MLLSDRDLRAEITAGRFSIDPFDDT
MAV_4828|M.avium_104 -------------------------MLLSDRDLRAEITAGRLGIEPFDDA
MMAR_0600|M.marinum_M -------------------------MLLSDRDLRAEISAGRLGIDPFDDS
MUL_0560|M.ulcerans_Agy99 -------------------------MLLSDRDLRAEISAGRLGIDPFDDS
:******:***: : *:.::*:* :
MSMEG_0678|M.smegmatis_MC2_155 LVQPSSIDVRLDRMFRVFNNTRYTHIDPAMQQDELTTLVEPAEGEPFVLH
TH_1210|M.thermoresistible__bu LVQPSSIDVRLDNLFRVFNNTRYTHIDPSIRQDELTSLVQAEDGEPFVLH
MAB_4297c|M.abscessus_ATCC_199 LIQPSSIDVRLDSLFRVFNNTRYTHIDPKERQDELTSLVEPAEGEPFVLH
Mflv_0291|M.gilvum_PYR-GCK LIQPSSVDVRLDNLFRVFNNTRYTHIDPAQRQDDLTSLVEPKEGEPFVLH
Mvan_0589|M.vanbaalenii_PYR-1 LVQPSSVDVRLDNLFRVFNNTRYTHIDPAQRQDDLTSLVEPKEGEPFVLH
Rv0321|M.tuberculosis_H37Rv LVQPSSIDVRLDCLFRVFNNTRYTHIDPAKQQDELTSLVQPVDGEPFVLH
Mb0329|M.bovis_AF2122/97 LVQPSSIDVRLDCLFRVFNNTRYTHIDPAKQQDELTSLVQPVDGEPFVLH
MLBr_02507|M.leprae_Br4923 LVQPSSIDVRLDCMFRVFNNTRYTHIDPARQQDELTSLVELVDGEPFVLH
MAV_4828|M.avium_104 LVQPSSIDVRLDCMFRVFNNTRYTHIDPAKQQDELTSLVEPQDGEPFVLH
MMAR_0600|M.marinum_M LVQPSSVDVRLDCMFRVFNNTRYTHIDPAQRQDELTSVVEPTKGDPFVLH
MUL_0560|M.ulcerans_Agy99 LVQPSSVDVRLDCMFRVFNNTRYTHIDPAQRQDELTSAVEPTKGDPFVLH
*:****:***** :************** :**:**: *: .*:*****
MSMEG_0678|M.smegmatis_MC2_155 PGEFVLGSTLELCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
TH_1210|M.thermoresistible__bu PGEFVLGSTLELCSLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
MAB_4297c|M.abscessus_ATCC_199 PGEFVLGSTLEVCSLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
Mflv_0291|M.gilvum_PYR-GCK PGEFVLGATLERCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
Mvan_0589|M.vanbaalenii_PYR-1 PGEFVLGATLERCTLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
Rv0321|M.tuberculosis_H37Rv PGEFVLGSTLELFTLPDNLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
Mb0329|M.bovis_AF2122/97 PGEFVLGSTLELFTLPDNLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
MLBr_02507|M.leprae_Br4923 PGGFVLGSTLELFTLPEDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFCGH
MAV_4828|M.avium_104 PGEFVLGSTLELITLPDDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
MMAR_0600|M.marinum_M PGEFVLGSTLEIFSLPGDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
MUL_0560|M.ulcerans_Agy99 PGEFVLGSTLEIFSLPGDLAGRLEGKSSLGRLGLLTHSTAGFIDPGFSGH
** ****:*** :** :*****************************.**
MSMEG_0678|M.smegmatis_MC2_155 ITLELSNVANLPITLWPGMKIGQLCLLRLTSPAENPYGSAAVGSKYQGQR
TH_1210|M.thermoresistible__bu ITLELSNVANLPITLWPGMKIGQLCLLRLTSPAEFPYGSTQAGSKYQGQR
MAB_4297c|M.abscessus_ATCC_199 ITLELSNVANLPITLWPGMKIGQLCLLRLTSPAEHPYGSAGVGSKYQGQR
Mflv_0291|M.gilvum_PYR-GCK ITLELSNVANLPITLWPGMKIGQLCLLRLTSPAQHPYGSSQVGSKYQGQR
Mvan_0589|M.vanbaalenii_PYR-1 ITLELSNVANLPITLWPGMKIGQLCLLRLTSPAEHPYGSSSVGSKYQGQR
Rv0321|M.tuberculosis_H37Rv ITLELSNVANLPITLWPGMKIGQLCMLRLTSPSEHPYGSSRAGSKYQGQR
Mb0329|M.bovis_AF2122/97 ITLELSNVANLPITLWPGMKIGQLCMLRLTSPSEHPYGSSRAGSKYQGQR
MLBr_02507|M.leprae_Br4923 ITLELSNVANLPITLWPGMKIGQLCVLRLTSPAEHPYGSASAGSKYQGQR
MAV_4828|M.avium_104 ITLELSNVANLPITLWPGMKIGQLCILRLTSPAEHPYGSTRVGSKYQGQR
MMAR_0600|M.marinum_M ITLELSNVANLPITLWPGMKIGQLCILKLTSPSEHPYGSSGVGSKYQGQR
MUL_0560|M.ulcerans_Agy99 ITLELSNVANLPITLWPGMKIGQLCILKLTSPSEHPYGSSGVGSKYQGQR
*************************:*:****:: ****: .********
MSMEG_0678|M.smegmatis_MC2_155 GPTPSRSHLNFIKS-----
TH_1210|M.thermoresistible__bu GPTPSRSYQNFVKSIRPQP
MAB_4297c|M.abscessus_ATCC_199 GPTPSRSYQNFINP-----
Mflv_0291|M.gilvum_PYR-GCK GPTPSKSYQNFVKSN----
Mvan_0589|M.vanbaalenii_PYR-1 GPTPSRSYQNFVKND----
Rv0321|M.tuberculosis_H37Rv GPTPSRSYQNFIRST----
Mb0329|M.bovis_AF2122/97 GPTPSRSCQNFIRST----
MLBr_02507|M.leprae_Br4923 GPTPSRSYENFIKNT----
MAV_4828|M.avium_104 GPTPSRSYQNFITST----
MMAR_0600|M.marinum_M GPTPSRSYQNFIRST----
MUL_0560|M.ulcerans_Agy99 GPTPSRSYQNFIRST----
*****:* **: