For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVLSLGIFLGPVAEADNPLDGQGFYVNPNSAAMRAAQGAGSPELTSIANTPQAYWMDNVSSPSVDAKYIS AAQASGATPVLALYAIPHRDCGSFAAGGFGSADAYKGWIDGVAAAIGAGPAVVILEPDALAMADCLSGAQ RQERFDLMRYAVDTLTRNPATALYIDGGHSRWVSADVMAARLNEVGVSKARGFSLNTANFFTTDEEIGYG DAISGMTNGAHYVIDTSRNGAGPTSDALYWCNPSGRALGTPPTTATASPNADAYLWVKRPGESDGSCDRG DPRAGTFVNQFAIDLARNAGR*
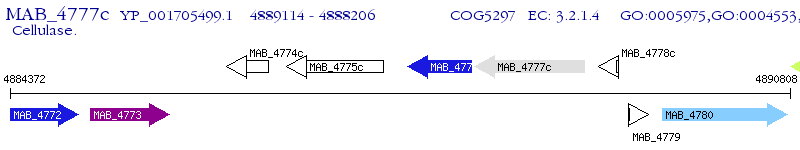
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4777c | - | - | 100% (302) | cellulase CelA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0063 | celA1 | 1e-122 | 66.35% (318) | endo-1,4-beta-glucanase |
| M. gilvum PYR-GCK | Mflv_1003 | - | 1e-122 | 72.98% (285) | cellulase |
| M. tuberculosis H37Rv | Rv0062 | celA1 | 1e-122 | 66.35% (318) | endo-1,4-beta-glucanase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0107 | celA | 1e-114 | 65.99% (297) | cellobiohydrolase a (1,4-beta-cellobiosidase a) CelA |
| M. avium 104 | MAV_0326 | - | 1e-122 | 69.74% (304) | endoglucanase A |
| M. smegmatis MC2 155 | MSMEG_6752 | - | 1e-130 | 77.59% (290) | endoglucanase A |
| M. thermoresistible (build 8) | TH_0192 | celA | 1e-123 | 74.65% (284) | POSSIBLE CELLULASE CELA1 (ENDOGLUCANASE) |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5837 | - | 1e-119 | 71.03% (290) | cellulase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1003|M.gilvum_PYR-GCK -------------------------------------------MSSAARA
Mvan_5837|M.vanbaalenii_PYR-1 --------------------------------MFGGTAKFPQVTFSAAGV
TH_0192|M.thermoresistible__bu ----------------------------------------VDVMSTAAHA
MSMEG_6752|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4777c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0063|M.bovis_AF2122/97 MTRRTGQRWRGTLPGRRPWTRPAPATCRRHLAFVELRHYFARVMSSAIGS
Rv0062|M.tuberculosis_H37Rv MTRRTGQRWRGTLPGRRPWTRPAPATCRRHLAFVELRHYFARVMSSAIGS
MMAR_0107|M.marinum_M ------------------------------MAFVELWHYFAGVMSSAFGA
MAV_0326|M.avium_104 --------------------------------------------------
Mflv_1003|M.gilvum_PYR-GCK AARWIAPFLTVAAVAGFAVAAQPAHRGTAPEVRLVSDANPLVGRPFYVNP
Mvan_5837|M.vanbaalenii_PYR-1 IARWITPCLAVAAIAATGVVAEPGPIPAAPVVRLASEANPLVGLPFYVNP
TH_0192|M.thermoresistible__bu AARSILPLLTVAAVA-VGVLTGTAP-GTTPAIRLVSDANPLAGHTFYVNP
MSMEG_6752|M.smegmatis_MC2_155 --------MAVTALAGAGALAEPVRPGAAIEVRLASDGNPLAGQTFYVDP
MAB_4777c|M.abscessus_ATCC_199 -----------MAVLSLGIFLGP----------VAEADNPLDGQGFYVNP
Mb0063|M.bovis_AF2122/97 VARWIVPLLGVAAVASIGVIADPVRVVRAPALILVDAANPLAGKPFYVDP
Rv0062|M.tuberculosis_H37Rv VARWIVPLLGVAAVASIGVIADPVRVVRAPALILVDAANPLAGKPFYVDP
MMAR_0107|M.marinum_M LARRIAPFLAVSAAAYLAFFAAPAPVVPAPPITLVDGANPLAGQPFYVDP
MAV_0326|M.avium_104 ----------------MGPVAEP-----APAIRLAADGNPLAGAPFYVNP
. . :. *** * ***:*
Mflv_1003|M.gilvum_PYR-GCK NSKGTRAAQSN--PDPLLAAVVNTPTAYWMDH-ISTPAVD---AKYIATA
Mvan_5837|M.vanbaalenii_PYR-1 QSKGMRAAQGN--PDPLLSAVVNTPTAYWMDH-ISTPAVD---AKYIATA
TH_0192|M.thermoresistible__bu DSKAARAAKGD--PSPELDIIVNTPTAYWMDH-LSSPAVD---AKYIADA
MSMEG_6752|M.smegmatis_MC2_155 NSKAMRALKGN--PSPELEMIANTPHAYWMDN-VSTFAVD---AKYIQTA
MAB_4777c|M.abscessus_ATCC_199 NSAAMRAAQGA--GSPELTSIANTPQAYWMDN-VSSPSVD---AKYISAA
Mb0063|M.bovis_AF2122/97 ASAAMIAARNANPPNAELTSVANTPQSYWLDQAFPPATVGGTVARYTGAA
Rv0062|M.tuberculosis_H37Rv ASAAMVAARNANPPNAELTSVANTPQSYWLDQAFPPATVGGTVARYTGAA
MMAR_0107|M.marinum_M GSAAMVAARNATPPSPELNAIANTPQSYWLDQAFPASTVGGTVARQTGAA
MAV_0326|M.avium_104 NSAAMRAAQSADPPSPELTTIANTPQAYWIVPGGSAATVG----KYVGDA
* . * :. .. * :.*** :**: .. :*. : *
Mflv_1003|M.gilvum_PYR-GCK QAAGTTPVLALYGIPNRDCGSYAAGGFGSAGSYRAWIDGVAGAIGGGPAA
Mvan_5837|M.vanbaalenii_PYR-1 QAAGTMPVLALYGIPNRDCGSYAAGGFGSAGAYRAWIDGVAAAIGPGRAA
TH_0192|M.thermoresistible__bu QAAGEMPILALYGIPHRDCGSYAAGGFGSAAAYRGWIDGVAAAIGSGPAA
MSMEG_6752|M.smegmatis_MC2_155 QAAGTMPILALYGIPNRDCGSLAAGGFGSAAAYRGWIDGVAGAIGSGPAA
MAB_4777c|M.abscessus_ATCC_199 QASGATPVLALYAIPHRDCGSFAAGGFGSADAYKGWIDGVAAAIGAGPAV
Mb0063|M.bovis_AF2122/97 QAAGAMPVLTLYGIPHRDCGSYASGGFATGTDYRGWIDAVASGLGSSPAT
Rv0062|M.tuberculosis_H37Rv QAAGAMPVLTLYGIPHRDCGSYASGGFATGTDYRGWIDAVASGLGSSPAT
MMAR_0107|M.marinum_M QAAGAMPILTLYGIPHRDCGSYASGGFATGADYRAWIDAISSGLGSAPAA
MAV_0326|M.avium_104 NAAGAIPVLAIYGIPHRDCGSFAAGGMASADDYRGWIDGIASGVGASRVA
:*:* *:*::*.**:***** *:**:.:. *:.***.::..:* . ..
Mflv_1003|M.gilvum_PYR-GCK VILEPDALAMIDCLSPGRQQERLELIGYAVDTLTRNPATAVYVDAGHSRW
Mvan_5837|M.vanbaalenii_PYR-1 VILEPDALAMIDCLSPGQQQERLELIGYAVDTLSRNPGTAVYVDAGHPRW
TH_0192|M.thermoresistible__bu VILEPDALAMSGCLSSEQRQERFELIRYAVDTLTRNPATAVYIDAGHPRW
MSMEG_6752|M.smegmatis_MC2_155 VILEPDALAMADCLSPDQRQERFDLIRYAVDTLTRNPATAVYVDAGHSRW
MAB_4777c|M.abscessus_ATCC_199 VILEPDALAMADCLSGAQRQERFDLMRYAVDTLTRNPATALYIDGGHSRW
Mb0063|M.bovis_AF2122/97 IIVEPDALAMADCLSPDQRQERFDLVRYAVDTLTRDPAAAVYVDAGHSRW
Rv0062|M.tuberculosis_H37Rv IIVEPDALAMADCLSPDQRQERFDLVRYAVDTLTRDPAAAVYVDAGHSRW
MMAR_0107|M.marinum_M IIVEPDALAMADCLSADQRQERFDLVRYAVNTLTRDPAAAVYVDGGHSRW
MAV_0326|M.avium_104 IVVEPDALAMADCLSGDQRQERFDLVRYAVDTLTRDPNAAVYVDAGHLRW
:::******* .*** ::***::*: ***:**:*:* :*:*:*.** **
Mflv_1003|M.gilvum_PYR-GCK VPADVMAGRLNQVGIAKARGFSLNTANFFTTEESVGYGGAISGMTGGKPF
Mvan_5837|M.vanbaalenii_PYR-1 VAADVMAGRLNQVGIDRARGFSLNTANFFTTEESMGYGGAISGMTGGKPF
TH_0192|M.thermoresistible__bu VSVEDMAGMLNEVGVARARGFSLNTANFFTTEESIGYGEAISGLTGGAHY
MSMEG_6752|M.smegmatis_MC2_155 VNAEDMAARLNEVGVTKTRGFSLNTANFFTTEEEIGYGEAISGLTNGSHY
MAB_4777c|M.abscessus_ATCC_199 VSADVMAARLNEVGVSKARGFSLNTANFFTTDEEIGYGDAISGMTNGAHY
Mb0063|M.bovis_AF2122/97 LSAEAMAARLNDVGVGRARGFSLNVSNFYTTDEEIGYGEAISGLTNGSHY
Rv0062|M.tuberculosis_H37Rv LSAEAMAARLNDVGVGRARGFSLNVSNFYTTDEEIGYGEAISGLTNGSHY
MMAR_0107|M.marinum_M VSAEEMAARLNQVDVGHARGFSLNISNFYTTDEEIGYGEAISGMTNGSHY
MAV_0326|M.avium_104 HSPEDMAARLNQAGVAHARGFSVNTANFYTTEDEIGYGEAISGLTNGAHY
: **. **:..: ::****:* :**:**::.:*** ****:*.* :
Mflv_1003|M.gilvum_PYR-GCK VIDTSRNGAGPVEGDDLYWCNPSGRALGVRPTTDTGNPMVDAFLWVKRPG
Mvan_5837|M.vanbaalenii_PYR-1 VIDTSRNGAGPVEGDDLYWCNPSGRALGARPTTNTGNPMVDAFLWVKRPG
TH_0192|M.thermoresistible__bu VIDTSRNGAGPLDSD--AWCNPPGRALGAFPTTATAGAHADAYLWIKRPG
MSMEG_6752|M.smegmatis_MC2_155 VIDTSRNGVGPVDSD--SWCNPPGRALGSPPTTATAGAHADAYLWVKRPG
MAB_4777c|M.abscessus_ATCC_199 VIDTSRNGAGP-TSDALYWCNPSGRALGTPPTTATASPNADAYLWVKRPG
Mb0063|M.bovis_AF2122/97 VIDTSRNGAGPAPDAPLNWCNPSGRALGAPPTTATAGAHADAYLWIKRPG
Rv0062|M.tuberculosis_H37Rv VIDTSRNGAGPAPDAPLNWCNPSGRALGAPPTTATAGAHADAYLWIKRPG
MMAR_0107|M.marinum_M VIDTGRNGVGPAPESPLNWCNPDGRALGVAPTTATAGAHADAYLWIKRPG
MAV_0326|M.avium_104 VIDTSRNGAGPAPDSDLNWCNPSGRALGTPPTAATAGAHADAYLWIKRPG
****.***.** **** ***** **: *... .**:**:****
Mflv_1003|M.gilvum_PYR-GCK ESDGACR-GAPSAGTFVAQYAIDLARNAGW
Mvan_5837|M.vanbaalenii_PYR-1 ESDGACR-GFPSAGTFMSQNAIDLARNAGW
TH_0192|M.thermoresistible__bu ESDGACN-GHPPAGTFVARYAVELVRNAGL
MSMEG_6752|M.smegmatis_MC2_155 ESDGACN-GHPSAGTFVSQYAIDLARNAGR
MAB_4777c|M.abscessus_ATCC_199 ESDGSCDRGDPRAGTFVNQFAIDLARNAGR
Mb0063|M.bovis_AF2122/97 ESDGTCGRGEPQAGRFVSQYAIDLAHNAGQ
Rv0062|M.tuberculosis_H37Rv ESDGTCGRGEPQAGRFVSQYAIDLAHNAGQ
MMAR_0107|M.marinum_M ESDGSCGRGEPQAGHFVSQYAIDLAHNAGQ
MAV_0326|M.avium_104 ESDGSCGKGDPPAGNFVNQYAIDLVHNVGH
****:* * * ** *: : *::*.:*.*