For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSVWRQESAHPGWPMLLGKLRVPAGVVSVRPIRLRDGMAWSRARLADQSHLQPWEPSSEIDWMARHGVSS WPALCSGLRAEARKGRMIPCAIELNGQFVGQLTIGNIAHGALRSAWIGYWVVSTVTRQGVATAALALGLD HCFGPVTLHRVEATVRPENGASRAVLSKVGFREEGLLKRYLDVDGAWRDHLLVAMTVEEVRGSVAATLVR SGNASWV
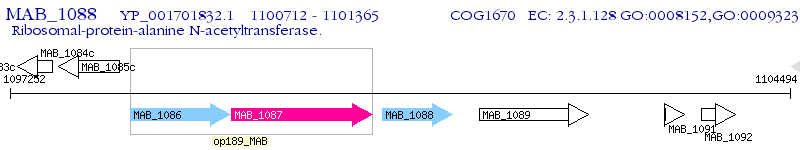
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1088 | - | - | 100% (217) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. abscessus ATCC 19977 | MAB_4621c | - | 1e-05 | 30.95% (84) | putative acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1022 | rimJ | 4e-89 | 74.26% (202) | ribosomal-protein-alanine acetyltransferase |
| M. gilvum PYR-GCK | Mflv_1899 | - | 5e-94 | 73.15% (216) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv0995 | rimJ | 4e-89 | 74.26% (202) | ribosomal-protein-alanine acetyltransferase |
| M. leprae Br4923 | MLBr_00184 | rimJ | 8e-91 | 71.36% (213) | putative acetyltransferase |
| M. marinum M | MMAR_4519 | rimJ | 3e-92 | 71.16% (215) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. avium 104 | MAV_1104 | - | 7e-97 | 75.12% (217) | acetyltransferase, GNAT family protein |
| M. smegmatis MC2 155 | MSMEG_5469 | - | 3e-88 | 71.50% (207) | acetyltransferase, GNAT family protein |
| M. thermoresistible (build 8) | TH_0938 | rimJ | 4e-92 | 70.64% (218) | POSSIBLE RIBOSOMAL-PROTEIN-ALANINE ACETYLTRANSFERASE RIMJ |
| M. ulcerans Agy99 | MUL_4691 | rimJ | 2e-87 | 73.13% (201) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. vanbaalenii PYR-1 | Mvan_4831 | - | 3e-96 | 73.61% (216) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1022|M.bovis_AF2122/97 ------------------MAVGPLRVSAGVIRLRPVRMRDGVHWSRIRLA
Rv0995|M.tuberculosis_H37Rv ------------------MAVGPLRVSAGVIRLRPVRMRDGVHWSRIRLA
MMAR_4519|M.marinum_M ----MNLMRAGQRHPGWPMDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLA
MUL_4691|M.ulcerans_Agy99 ------------------MDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLA
MLBr_00184|M.leprae_Br4923 -------MRFNARHPGWPSNVGPLRVPAGVIRLRAVRLRDGVQWSRIRLA
MAV_1104|M.avium_104 ----MNLLRNNSRHPGWPMSVGPLRVPAGVVRLRAVRLRDGAQWSRIRLA
Mflv_1899|M.gilvum_PYR-GCK ----MDLLSSRSQHPGWPAPVGPLRVSAGLVRLRPVRLRDAAQWSRIRLA
Mvan_4831|M.vanbaalenii_PYR-1 MAAVMNLLSSRSQHPGWPLAVGPLRVPAGVVRLRPVRLRDAAQWSRIRLA
MSMEG_5469|M.smegmatis_MC2_155 ------------MHPGWPDTVGPLRVPAGVVGLRPVRMRDAAAWSRIRLA
TH_0938|M.thermoresistible__bu ----VNLFRSSAAHPGWPIPAGPLRVKAGVIGLRPIRMRDAAVWSRLRLA
MAB_1088|M.abscessus_ATCC_1997 ----MSVWRQESAHPGWPMLLGKLRVPAGVVSVRPIRLRDGMAWSRARLA
* *** **:: :*.:*:**. *** ***
Mb1022|M.bovis_AF2122/97 DRAHLEPWEPSADGEWTVRHTVAAWPAVCSGLRSEARNGRMLPYVIELDG
Rv0995|M.tuberculosis_H37Rv DRAHLEPWEPSADGEWTVRHTVAAWPAVCSGLRSEARNGRMLPYVIELDG
MMAR_4519|M.marinum_M DRAHLEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDG
MUL_4691|M.ulcerans_Agy99 DRAHLEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDG
MLBr_00184|M.leprae_Br4923 DRAYLEPWEPSTEGDWVVRHSVIAWQVLCSSLRSEARKGRMLPYAIELDG
MAV_1104|M.avium_104 DRAHLEPWEPSAEGDWTTRHAVAAWPALCSGLRAEARQGRMLPYVIELDG
Mflv_1899|M.gilvum_PYR-GCK DRDHLEPWEPSTDMNWELRHSVSAWPSVCSGLRSEARKGRMLPYVIELDG
Mvan_4831|M.vanbaalenii_PYR-1 DRGHLEPWEPSTDMDWELRHSVSAWPSVCSGLRSEARKGRMLPYVIELDG
MSMEG_5469|M.smegmatis_MC2_155 DQHHLEPWEPMTGMDWKVRHAVTSWPSICSGLRAEARHGRMLPFVIELDG
TH_0938|M.thermoresistible__bu DRAHLEPWEPATGMDWETRHAISSWPSICSGLRAEARKGRLLPYVIELDG
MAB_1088|M.abscessus_ATCC_1997 DQSHLQPWEPSSEIDWMARHGVSSWPALCSGLRAEARKGRMIPCAIELNG
*: :*:**** : :* ** : :* :**.**:***:**::* .***:*
Mb1022|M.bovis_AF2122/97 QFCGQLTIGNVTHGALRSAWIGYWVPSAATGGGVATGALALGLDHCFGPV
Rv0995|M.tuberculosis_H37Rv QFCGQLTIGNVTHGALRSAWIGYWVPSAATGGGVATGALALGLDHCFGPV
MMAR_4519|M.marinum_M RFCGQLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPV
MUL_4691|M.ulcerans_Agy99 RFCGQLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPV
MLBr_00184|M.leprae_Br4923 NFCGQLTIGNVTHGALRSAWIGYWVSSSATGGGVATGALALGLDHCFGPV
MAV_1104|M.avium_104 RFCGQLTIGNVTHGALRSAWIGYWVPRSATGGGVATAALALGLDHCFGPV
Mflv_1899|M.gilvum_PYR-GCK QFAGQLTIGNVTHGALRSAWIGYWVASGSTGGGVATAALALGLDHCFGPV
Mvan_4831|M.vanbaalenii_PYR-1 QFAGQLTIGNVTHGALRSAWIGYWVASGSTGGGVATAALALGLDHCFGPV
MSMEG_5469|M.smegmatis_MC2_155 EFVGQLTIGNVTHGALRSAWIGYWVASSRTGGGIATAALAMGLDHCFTAV
TH_0938|M.thermoresistible__bu RFVGQLTIGNVTHGALRSAWIGYWVASAVTGGGVATGALALGVDHCFSRV
MAB_1088|M.abscessus_ATCC_1997 QFVGQLTIGNIAHGALRSAWIGYWVVSTVTRQGVATAALALGLDHCFGPV
.* ****:**:.************* * *:**.*:*:*:**** *
Mb1022|M.bovis_AF2122/97 MLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMAIT
Rv0995|M.tuberculosis_H37Rv MLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMAIT
MMAR_4519|M.marinum_M GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLT
MUL_4691|M.ulcerans_Agy99 GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLT
MLBr_00184|M.leprae_Br4923 MLHRVEATVRPANVASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMALT
MAV_1104|M.avium_104 MLHRVEATVRPENAASRAVLAKVGFRQEGLLQRYLEVDKAWRDHLLMAIT
Mflv_1899|M.gilvum_PYR-GCK MLHRVEATVRPENGPSRAVLAKAGLREEGLLKRYLDVDGAWRDHLLVAIT
Mvan_4831|M.vanbaalenii_PYR-1 MLHRVEATVRPENGPSRAVLAKAGFREEGLLKRYLDVDGAWRDHLLVAIT
MSMEG_5469|M.smegmatis_MC2_155 QLHRIEATVRPENTPSRAVLAHVGFREEGLLKRYLEVDGAWRDHLLVAIT
TH_0938|M.thermoresistible__bu GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLDVDGAWRDHLLVAIT
MAB_1088|M.abscessus_ATCC_1997 TLHRVEATVRPENGASRAVLSKVGFREEGLLKRYLDVDGAWRDHLLVAMT
***:****** * .*****::.*:*:****:***:** *******:.:*
Mb1022|M.bovis_AF2122/97 VEEVYG-SVASTLVRAGHASWP
Rv0995|M.tuberculosis_H37Rv VEEVYG-SVASTLVRAGHASWP
MMAR_4519|M.marinum_M VEEVEG-SVVSTLVRAGQASRL
MUL_4691|M.ulcerans_Agy99 VEEVEG-SVVSTLVRAGQASRL
MLBr_00184|M.leprae_Br4923 IEEVSG-SVTSNLVRAGRASWL
MAV_1104|M.avium_104 VEEVSG-SVASTLVRAGNASWV
Mflv_1899|M.gilvum_PYR-GCK AEEVNG-SVASALVRAGHASWA
Mvan_4831|M.vanbaalenii_PYR-1 IEELNG-TAASALVRAGRASWA
MSMEG_5469|M.smegmatis_MC2_155 AEELPQ-SAAHRLVAAGRAEWC
TH_0938|M.thermoresistible__bu VEELTSGSATAALVKHGRAEWV
MAB_1088|M.abscessus_ATCC_1997 VEEVRG-SVAATLVRSGNASWV
**: :.. ** *.*.