For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRCDGQERGCAEAGSATDELPGTVGAASSGRTDSQNHHPRRVSSFRSRGSSLSEGQQKAWDRSWPTYGRL AREADGSCPPLDPATWFGRTAPLILEIGCGTGTSTAAMALQEPDFDVLAVEVYRKGLAQLLSAIDREGIE NIRLIRGDALDVLEHLLPSGSLTAARVFFPDPWPKARHHKRRLLQPGTVALLSDRLRPGGTLHVATDHAN YAEHIAEVGDGEPTLRRLRLDDPRIPVSVNRPTTKFEGKAHTVGSVINDFVWERL
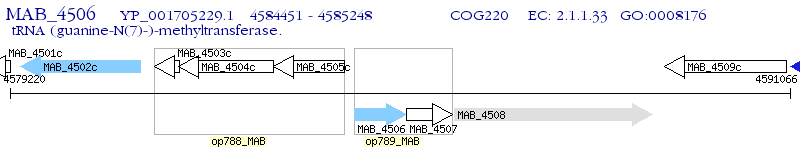
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4506 | - | - | 100% (265) | tRNA (guanine-N(7)-)-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0214c | trmB | 5e-70 | 56.70% (224) | tRNA (guanine-N(7)-)-methyltransferase |
| M. gilvum PYR-GCK | Mflv_0454 | trmB | 2e-84 | 61.85% (249) | tRNA (guanine-N(7))-methyltransferase |
| M. tuberculosis H37Rv | Rv0208c | trmB | 5e-70 | 56.70% (224) | tRNA (guanine-N(7)-)-methyltransferase |
| M. leprae Br4923 | MLBr_02622 | trmB | 1e-70 | 55.41% (222) | tRNA (guanine-N(7)-)-methyltransferase |
| M. marinum M | MMAR_0448 | trmB | 4e-74 | 59.74% (231) | S-adenosylmethionine-dependentmethyltransferase |
| M. avium 104 | MAV_4966 | trmB | 1e-81 | 61.09% (239) | tRNA (guanine-N(7))-methyltransferase |
| M. smegmatis MC2 155 | MSMEG_0252 | trmB | 2e-81 | 62.18% (238) | tRNA (guanine-N(7))-methyltransferase |
| M. thermoresistible (build 8) | TH_0722 | - | 1e-83 | 65.18% (224) | HYPOTHETICAL METHLYTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_1098 | trmB | 6e-74 | 59.31% (231) | tRNA (guanine-N(7)-)-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0198 | trmB | 5e-82 | 60.73% (247) | tRNA (guanine-N(7))-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0448|M.marinum_M MPAATTRRQLLGINLDQERLRPGFMRHDGPMHVQPGVGLQSDTSSSTGTG
MUL_1098|M.ulcerans_Agy99 ------------------------MRHDGPMHVQPGVGLQSDTSSSTGTG
Mb0214c|M.bovis_AF2122/97 ------------------------MVHHGQMHAQPGVGLRPDTPVAS---
Rv0208c|M.tuberculosis_H37Rv ------------------------MVHHGQMHAQPGVGLRPDTPVAS---
MLBr_02622|M.leprae_Br4923 --------------------------------------------------
MAV_4966|M.avium_104 ------------------------MGHHGQMHAQHGVTMPADSPTGQ---
Mflv_0454|M.gilvum_PYR-GCK ------------------------MSDHGRMHSTGSEVAAPVAPDPDTEG
Mvan_0198|M.vanbaalenii_PYR-1 ------------------------MSDHGRMHIPESGLATPAAAHSDDP-
MSMEG_0252|M.smegmatis_MC2_155 ------------------------------MNAS----AGPPA-------
TH_0722|M.thermoresistible__bu --------------------------------------------------
MAB_4506|M.abscessus_ATCC_1997 ------------------------MRCDGQERGCAEAGSATDELPGTVGA
MMAR_0448|M.marinum_M SGPADEPEAEKSAWGYLPPTAFRSRHSALSSIQQQTWERLWPQLGRQATA
MUL_1098|M.ulcerans_Agy99 SGPADEPEAEKSAWGYLPPTAFRSRHSALSSIQQQTWERRWPQLGRQATA
Mb0214c|M.bovis_AF2122/97 --------------GQLPSTSIRSRRSGISKAQRETWERLWPELGLLALP
Rv0208c|M.tuberculosis_H37Rv --------------GQLPSTSIRSRRSGISKAQRETWERLWPELGLLALP
MLBr_02622|M.leprae_Br4923 ------------MARHLPATAFRKRRSALSHAQRQTWERLWPEIGIS---
MAV_4966|M.avium_104 --------PDRSEQRYFPATAFRTRRSTLSDAQRQTWDRRWPELGLSVGP
Mflv_0454|M.gilvum_PYR-GCK ----------VHPHFNRRVTSFRARRSTISDGQQATWDRLWPQLGRQARD
Mvan_0198|M.vanbaalenii_PYR-1 ----------PHPHFNRRVTSFRARRSTISDAQQATWDRLWPELGRQVRD
MSMEG_0252|M.smegmatis_MC2_155 -----------EPHRLPRVTSFRTRRSTLSGGQQGTWDRLWPKLGMQARD
TH_0722|M.thermoresistible__bu ------------------VTSFRTRRTTLSRGQQETWERLWPVLGRQARD
MAB_4506|M.abscessus_ATCC_1997 ASSG---RTDSQNHHPRRVSSFRSRGSSLSEGQQKAWDRSWPTYGRLARE
:::* * : :* *: :*:* ** *
MMAR_0448|M.marinum_M HSQPSARGHE-PIDPIDARAWFGREAPVVLEIGCGSGTSTLAMAQAEPNV
MUL_1098|M.ulcerans_Agy99 HSQPSARGHE-PIDPIDARAWFGREAPVVLEIGCGSGTSTLAMAQAEPNV
Mb0214c|M.bovis_AF2122/97 QS---PRG-----TPVDTRAWFGRDAPVVLEIGSGSGTSTLAMAKAEPHV
Rv0208c|M.tuberculosis_H37Rv QS---PRG-----TPVDTRAWFGRDAPVVLEIGSGSGTSTLAMAKAEPHV
MLBr_02622|M.leprae_Br4923 AVSQTRSA-----ERLDTGAWFGRSTLVVLEVGCGSGTATLAMAQHEPDI
MAV_4966|M.avium_104 AEDPDRPG-----PPLDTDAWFGRKAPVVLEIGCGNGTSTLAMAKQEPGV
Mflv_0454|M.gilvum_PYR-GCK GDEPP--------GPLDTDAWFGRHAPVVLEIGCGTGTSTLAMAQAEPGI
Mvan_0198|M.vanbaalenii_PYR-1 GDAPAGRLDTTPAGRLDTEAWFGRHAPVVLEIGCGTGTSTLAMAQAEPDI
MSMEG_0252|M.smegmatis_MC2_155 EDSQP-------ITDLDTEAWFGRSAPLVLEIGSGTGTSTLEMAKAEPHI
TH_0722|M.thermoresistible__bu ADGRPAPR-------LDLEEWFGRSAPVVLEIGCGSGVSTLAMAQQEPHL
MAB_4506|M.abscessus_ATCC_1997 ADGSCP--------PLDPATWFGRTAPLILEIGCGTGTSTAAMALQEPDF
:* **** : ::**:*.*.*.:* ** ** .
MMAR_0448|M.marinum_M DVIAVEVYRRGLAQLLCAIDRA---DLQRINIRLIRGNGIDVLRDLIAPE
MUL_1098|M.ulcerans_Agy99 DVIAVEVYRRGLAQLLCAIDRA---DLQRINIRLIRGNGIDVLRDLIAPE
Mb0214c|M.bovis_AF2122/97 DVIAVDVYRRGLAQLLCAIDKV---GSDGINIRLILGNAVDVLQHLIAPD
Rv0208c|M.tuberculosis_H37Rv DVIAVDVYRRGLAQLLCAIDKV---GSDGINIRLILGNAVDVLQHLIAPD
MLBr_02622|M.leprae_Br4923 DVIAVEVYRRGLAQLLCAIDRD---QVH--NIRMIHGNALYVLQYLIAPR
MAV_4966|M.avium_104 DVIAVEVYRRGLAQLLCAIDRD---NVT--NIRLIRGNALDVLQRLIAPA
Mflv_0454|M.gilvum_PYR-GCK DVVAVEVYRRGLAQLLSAIDRTAATEHPVSNIRLIRGDGVDVLTHMFGSA
Mvan_0198|M.vanbaalenii_PYR-1 DVVAVEVYRRGLAQLLSAMDR-----EGVTNIRLIRGDGVDVLTHMFGPD
MSMEG_0252|M.smegmatis_MC2_155 DVIAVEVYRKGLAQLLSAIDR-----EGVTNIRMIRGDGVDVLEHMIGSN
TH_0722|M.thermoresistible__bu DFIAVEVYKRGLAQLLSAIDRA-----GVSNIRLIRGDGVDVLEHLLGPE
MAB_4506|M.abscessus_ATCC_1997 DVLAVEVYRKGLAQLLSAIDRE-----GIENIRLIRGDALDVLEHLLPSG
*.:**:**::******.*:*: ***:* *:.: ** :: .
MMAR_0448|M.marinum_M SLTGVRVFFPDPWPKARHHKRRLIQPSTVALIADRLLPGGVFHAATDHPG
MUL_1098|M.ulcerans_Agy99 SLTGVRVFFPDPWPKARHHKRRLIQPSTVVLIADRLLPGGVFHAATDHPG
Mb0214c|M.bovis_AF2122/97 SLCGVRVFFPDPWPKARHHKRRLLQPATMALIADRLVPSGVLHAATDHPG
Rv0208c|M.tuberculosis_H37Rv SLCGVRVFFPDPWPKARHHKRRLLQPATMALIADRLVPSGVLHAATDHPG
MLBr_02622|M.leprae_Br4923 SLTGVRVFFPDPWPKVRHHKRRFLQPATVELIADRLLPGGVLHTATDHPD
MAV_4966|M.avium_104 SLTGVRVFFPDPWPKARHHKRRFLQPGTVGLIADRLLPGGVLHVATDHAG
Mflv_0454|M.gilvum_PYR-GCK SLTGVRVFFPDPWPKARHHKRRLLQPDTMALIADRLRPGGVLHAATDHQG
Mvan_0198|M.vanbaalenii_PYR-1 SLTGVRVFFPDPWPKARHHKRRLLQPDTVALIADRLRPGGILHAATDHMG
MSMEG_0252|M.smegmatis_MC2_155 TLTGVRVFFPDPWPKARHHKRRLLQPATVALIADRLREGGVLHAATDHMG
TH_0722|M.thermoresistible__bu SLIGVRVFFPDPWPKARHHKRRLLQPATVALIADRLKPGGVLHVATDHAG
MAB_4506|M.abscessus_ATCC_1997 SLTAARVFFPDPWPKARHHKRRLLQPGTVALLSDRLRPGGTLHVATDHAN
:* ..**********.******::** *: *::*** .* :*.**** .
MMAR_0448|M.marinum_M YAEHIIAAGDAEPALARVDPGVDSLPVSVVRPTTKYEMKAHNAGSSINEL
MUL_1098|M.ulcerans_Agy99 YAEHIIAAGDAEPALARVDPGVDSLPVSVVRPTTKYEMKAHNAGSSINEL
Mb0214c|M.bovis_AF2122/97 YAEHIAAAGDAEPRLVRVDPDTELLPISVVRPATKYERKAQLGGGAVIEL
Rv0208c|M.tuberculosis_H37Rv YAEHIAAAGDAEPRLVRVDPDTELLPISVVRPATKYERKAQLGGGAVIEL
MLBr_02622|M.leprae_Br4923 YAKQIAKFGDGEPLLSRADGRTQ-LPISTVRPTTKYETKAQHAGNVVTEL
MAV_4966|M.avium_104 YAEHIADVGAGEPRLRPADPDSP-LPISVARPTTKYETKAQDAGSAVTEF
Mflv_0454|M.gilvum_PYR-GCK YAAHIAEVGDAEPRLRRVAMDSADLPMSVTRPVTKYERKA-LAGPVVTEL
Mvan_0198|M.vanbaalenii_PYR-1 YAGHIAEVGDAEPRLRRIDPDDAGLPISTARPVTKYQRKA-LAGTEVAEL
MSMEG_0252|M.smegmatis_MC2_155 YAEHIAEVGDAEPLLRRVS-TSDDLPISLERPVTKYQRKA-LAGTEVAEL
TH_0722|M.thermoresistible__bu YAEFIAETGDAEPRLERVTGDLSALPISVRRPVTKYEGKAREANRAVADL
MAB_4506|M.abscessus_ATCC_1997 YAEHIAEVGDGEPTLRRLRLDDPRIPVSVNRPTTKFEGKAHTVGSVINDF
** * * .** * :*:* **.**:: ** . : ::
MMAR_0448|M.marinum_M IWKKQR---------
MUL_1098|M.ulcerans_Agy99 IWKKQR---------
Mb0214c|M.bovis_AF2122/97 LWKKHGCSERDLKIR
Rv0208c|M.tuberculosis_H37Rv LWKKHGCSERDLKIR
MLBr_02622|M.leprae_Br4923 IWKKRS---------
MAV_4966|M.avium_104 IWLRR----------
Mflv_0454|M.gilvum_PYR-GCK LWERTP---------
Mvan_0198|M.vanbaalenii_PYR-1 LWEKTTP--------
MSMEG_0252|M.smegmatis_MC2_155 VWEKRL---------
TH_0722|M.thermoresistible__bu LWSRKA---------
MAB_4506|M.abscessus_ATCC_1997 VWERL----------
:* :