For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGGTKRLPRAVREQQMLDAAVQEFALNGFRDTSMDAIAAAAKISKPMLYLYYGSKEDLFVACLRRELGRF VEAVRGDIDLTQSPRDVLSTTVVSFLNYVDTHRASWIVIYGQATSTQLFAKQVREGRDRIIELVARLIKA ASPEIDPETDYDAMAVALVGGGEAVASRVATGDIKVESAKDTLLNLGWRGLRGRPRDEPVAKRA*
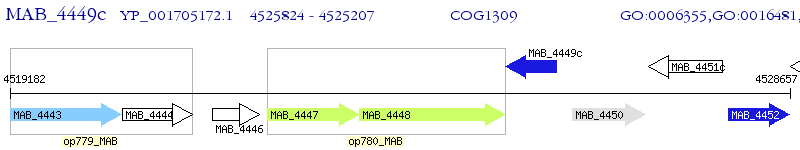
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4449c | - | - | 100% (205) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3199c | - | 2e-17 | 33.16% (196) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0476c | - | 6e-16 | 31.76% (170) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0244 | - | 1e-68 | 62.63% (198) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0414 | - | 6e-72 | 65.31% (196) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0238 | - | 1e-68 | 62.63% (198) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02568 | - | 5e-69 | 63.13% (198) | putative TetR-family transcriptional regulator |
| M. marinum M | MMAR_0500 | - | 4e-70 | 61.69% (201) | TetR family transcriptional regulator |
| M. avium 104 | MAV_4919 | - | 1e-71 | 64.85% (202) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0363 | - | 2e-73 | 66.16% (198) | TetR-family protein regulatory protein |
| M. thermoresistible (build 8) | TH_1911 | - | 2e-66 | 65.38% (182) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_1163 | - | 3e-70 | 61.69% (201) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0257 | - | 4e-71 | 64.29% (196) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0500|M.marinum_M --MAGGTKRLPRAIREQQMLDAAVQMFSANGYHETSMDTIAAAAQISKPM
MUL_1163|M.ulcerans_Agy99 --MAGGTKRLPRAIREQQMLDAAVQMFSANGYHETSMDTIAAAAQISKPM
Mb0244|M.bovis_AF2122/97 --MAGGTKRLPRAVREQQMLDAAVQMFSVNGYHETSMDAIAAEAQISKPM
Rv0238|M.tuberculosis_H37Rv --MAGGTKRLPRAVREQQMLDAAVQMFSVNGYHETSMDAIAAEAQISKPM
MLBr_02568|M.leprae_Br4923 --MAGGAKRIPRAVREQQMLDAAVQMFSVNGYHKTSMDAIAAEAQISKPM
MAV_4919|M.avium_104 --MAGGTKRLPRAVREQQMLDAAVQMFSVNGYHETSMDAIAAQAQISKPM
Mflv_0414|M.gilvum_PYR-GCK --MAGGTKRLPRAVREQQMLDAAVEIFSVNGYHETSMDAIAAEAQISKPM
Mvan_0257|M.vanbaalenii_PYR-1 --MAGGTKRLPRAVREQQMLDAAVEIFSVNGYHETSMDAIAAEAQISKPM
MSMEG_0363|M.smegmatis_MC2_155 MGMAGGTKRLPRAVREQQMLDAAVQMFSVNGYHETSMDAIAAAAEISKPM
TH_1911|M.thermoresistible__bu ------------------MLDAAVQMFSVNGYHETSMDAIAAQAEISKPM
MAB_4449c|M.abscessus_ATCC_199 --MAGGTKRLPRAVREQQMLDAAVQEFALNGFRDTSMDAIAAAAKISKPM
******: *: **::.****:*** *:*****
MMAR_0500|M.marinum_M LYLYYGSKEDLFGACLNREMSRFIDAVRADIDLSQSPKDLLRNAIVSFLR
MUL_1163|M.ulcerans_Agy99 LYLYYGSKEDLFGACLNREMSRFIDAVRADIDLSQSPKDLLRNAIVSFLR
Mb0244|M.bovis_AF2122/97 LYLYYGSKEDLFGACLNREMSRFIDALRSSINFDQSPKDLLRNTIVSFLR
Rv0238|M.tuberculosis_H37Rv LYLYYGSKEDLFGACLNREMSRFIDALRSSINFDQSPKDLLRNTIVSFLR
MLBr_02568|M.leprae_Br4923 LYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLRNTIVSFLR
MAV_4919|M.avium_104 LYLYYGSKEDLFGACLNRELGRFIDAVRADIDFQQSPNDMLRNTIASFMR
Mflv_0414|M.gilvum_PYR-GCK LYLYYGSKEELFGACLHRELTRFVDEVNSNIDFTAPPKELLRTAVLAFLN
Mvan_0257|M.vanbaalenii_PYR-1 LYLYYGSKEELFGACLNRELGRFVDEVNSNIDFSAAPKELLRTAVLAFLK
MSMEG_0363|M.smegmatis_MC2_155 LYLYYGSKEELFGACLDRELRRFVDTVRGQIDFSQSPKELLRTAVLAFLT
TH_1911|M.thermoresistible__bu LYLYYGSKEELFGACLKRELSRFVETVRADIDFSQSPKDLLRNAVRSFLT
MAB_4449c|M.abscessus_ATCC_199 LYLYYGSKEDLFVACLRRELGRFVEAVRGDIDLTQSPRDVLSTTVVSFLN
*********:** *** **: **: :...*:: .*.::* .:: :*:
MMAR_0500|M.marinum_M YIDQNQASWIVMYTQAVSSQAFAQTVREGREQIVELVAGMLRAGTRSPRS
MUL_1163|M.ulcerans_Agy99 YIDQNQASWIVMYTQAVSSQAFAQTVREGREQIVELVAGMLRAGTRSPRS
Mb0244|M.bovis_AF2122/97 YIDANRASWIVMYTQATSSQAFAHTVREGREQIVQLVAELVRAGTRGPLT
Rv0238|M.tuberculosis_H37Rv YIDANRASWIVMYTQATSSQAFAHTVREGREQIVQLVAELVRAGTRGPLT
MLBr_02568|M.leprae_Br4923 YIDANRASWIVMYIQAMSLPAFAQTVREARDQITELVAGLMRVGTRSSRP
MAV_4919|M.avium_104 YIDANRASWIVMYTQATSSQAFAHMVREGREQIIELVAGLMRAGSRTPRT
Mflv_0414|M.gilvum_PYR-GCK YIDANRASWMVLYTQATSSQAFAHTVREGRERIIEIVGRLLRSGTRFPEP
Mvan_0257|M.vanbaalenii_PYR-1 YIDANRASWMVLYTQATSSQAFAHTVREGRERIIELVGRLLRSGTKFPEP
MSMEG_0363|M.smegmatis_MC2_155 YIDRNRASWMVLYTQATSSQAFAHTVREGRERIIDLVARLLSTGTRNPQP
TH_1911|M.thermoresistible__bu YIDTNRASWLVLYTQATSSQAFAHTVREGRERIIDLVARLLRSGTRHPEP
MAB_4449c|M.abscessus_ATCC_199 YVDTHRASWIVIYGQATSTQLFAKQVREGRDRIIELVARLIKAASPEIDP
*:* ::***:*:* ** * **: ***.*::* ::*. :: .: .
MMAR_0500|M.marinum_M DAEIDMMAAALVGAGEAVANRLSAGDTDVDEAAEMMIDLFWRGLKGAPSD
MUL_1163|M.ulcerans_Agy99 DAEIDMMAAALVGAGEAVANRLSAGDTDVDEAAEMMIDLLWRGLKGAPSD
Mb0244|M.bovis_AF2122/97 DAEIEMMAVALVGAGEAVATRLGIGDTDVDEAAEMMINLFWLGLKGAPVD
Rv0238|M.tuberculosis_H37Rv DAEIEMMAVALVGAGEAVATRLGIGDTDVDEAAEMMINLFWLGLKGAPVD
MLBr_02568|M.leprae_Br4923 DHEYEIMAGALVGAGEAMATRLTTGDIAVDEAAELMINLFWHGLKGAPAD
MAV_4919|M.avium_104 DREHEMMAVALVGAGEAVANRLSTGDIDVDEAAALMIDLFWYGLKGSPEE
Mflv_0414|M.gilvum_PYR-GCK DTDFDMMAVALVGAGEAIASRVSTGDADVHEASELMINLFWRGLKGSPSA
Mvan_0257|M.vanbaalenii_PYR-1 DTDFELMAVALVGAGEAIASRVSTGDADVDEAAELMINLFWLGLKGAPSA
MSMEG_0363|M.smegmatis_MC2_155 DSDFEMMAVALVGAGEAIATRVSTGDADVDEAAELMINLFWRGLKGTPSD
TH_1911|M.thermoresistible__bu DTDFDMMAVALVGAGEAVATRVSAGDAEVDEAAELLINLFWRGLKGTPAE
MAB_4449c|M.abscessus_ATCC_199 ETDYDAMAVALVGGGEAVASRVATGDIKVESAKDTLLNLGWRGLRGRPRD
: : : ** ****.***:*.*: ** *..* :::* * **:* *
MMAR_0500|M.marinum_M RE----------IGSNFAAG-------
MUL_1163|M.ulcerans_Agy99 RE----------IGSNFAAG-------
Mb0244|M.bovis_AF2122/97 R-----------LETGH----------
Rv0238|M.tuberculosis_H37Rv R-----------LETGH----------
MLBr_02568|M.leprae_Br4923 RD----------FGPSIVAG-------
MAV_4919|M.avium_104 RE----------AAAAQPEADRSGAGG
Mflv_0414|M.gilvum_PYR-GCK ASDRDA----DATDATAVITS------
Mvan_0257|M.vanbaalenii_PYR-1 AD----------VDATAVISS------
MSMEG_0363|M.smegmatis_MC2_155 AG----------TPPVSAGSSG-----
TH_1911|M.thermoresistible__bu SHEDTHRENGQDVDRDGVDQSTTV---
MAB_4449c|M.abscessus_ATCC_199 EP----------VAKRA----------