For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YRLLKRFWIPLLLVVVVGIGGYAIAQIRGSAAPRPAKLGEGSGITANFNPKHITYEVVGSGGTANLNYLD ENGQPHLIENAPLPWSFTIVTTLPSMSANIVAQGDRSVTDLGCRVTVDGQVRDDRSSADKVKPFIYCLVK SV*
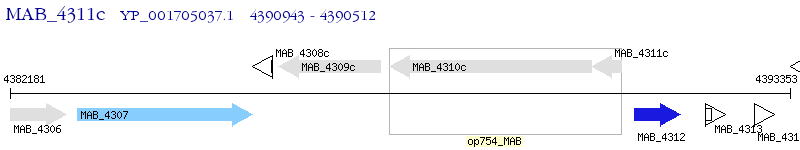
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4311c | - | - | 100% (143) | MmpS family protein |
| M. abscessus ATCC 19977 | MAB_3149 | - | 1e-29 | 42.07% (145) | MmpS family protein |
| M. abscessus ATCC 19977 | MAB_4383c | - | 1e-27 | 43.97% (141) | putative membrane protein, MmpS |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0696c | mmpS5 | 3e-28 | 45.89% (146) | hypothetical protein Mb0696c |
| M. gilvum PYR-GCK | Mflv_3028 | - | 1e-25 | 41.26% (143) | membrane family protein |
| M. tuberculosis H37Rv | Rv0677c | mmpS5 | 3e-28 | 45.89% (146) | hypothetical protein Rv0677c |
| M. leprae Br4923 | MLBr_02377 | - | 9e-28 | 40.69% (145) | hypothetical protein MLBr_02377 |
| M. marinum M | MMAR_3743 | mmpS5_1 | 3e-33 | 50.00% (144) | membrane protein MmpS5_1 |
| M. avium 104 | MAV_4756 | - | 1e-42 | 64.23% (137) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_0462 | - | 1e-61 | 74.83% (143) | MmpS4 protein |
| M. thermoresistible (build 8) | TH_2866 | - | 8e-31 | 45.39% (141) | PUTATIVE hypothetical protein Mvan_3834 |
| M. ulcerans Agy99 | MUL_3687 | mmpS5_1 | 1e-33 | 50.35% (141) | membrane protein MmpS5_1 |
| M. vanbaalenii PYR-1 | Mvan_1058 | - | 2e-28 | 44.68% (141) | hypothetical protein Mvan_1058 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3743|M.marinum_M ------MPTIPVTSVLKRAWIPLLMVVVLAISGLVVSRLHKIFGSEDLNA
MUL_3687|M.ulcerans_Agy99 --------------MLKRAWIPLLMVVVLAISGFVVSRLHKIFGSEDLNA
Mb0696c|M.bovis_AF2122/97 -----------MIGTLKRAWIPLLILVVVAIAGFTVQRIRTFFGSEGILV
Rv0677c|M.tuberculosis_H37Rv -----------MIGTLKRAWIPLLILVVVAIAGFTVQRIRTFFGSEGILV
Mflv_3028|M.gilvum_PYR-GCK -----------MITVVKRVWLPVLIVVALAIGFVSISYLRSVFGSNGAVV
Mvan_1058|M.vanbaalenii_PYR-1 -----------MITVVKRVWLPVLVIAAVAVGAVTVANLRSVFGSDKAIV
MLBr_02377|M.leprae_Br4923 MSKRQRGKGISIFKLLSRIWIPLVILVVLVVGGFVVYRVHSYFASEKRES
TH_2866|M.thermoresistible__bu -LWTSKAKDSAILTRLARAWIPLVMVVVVAIGGFAVWRIRGIFGSEQLPT
MAB_4311c|M.abscessus_ATCC_199 -----------MYRLLKRFWIPLLLVVVVGIGGYAIAQIRGSAAPR-PAK
MSMEG_0462|M.smegmatis_MC2_155 -------MGYAVYRVMKRFWIPLLLVVVVAVGAYAIVRIREAAGAH-APT
MAV_4756|M.avium_104 --------------------MPLLLVVVISLGAYAVVRIRDTFGTHGGVT
:*::::..: :. : :: ..
MMAR_3743|M.marinum_M NAGAGIDIVQ-FNPKVVIYEISGPAGSTANINFWDADANTHQVNAAPLPW
MUL_3687|M.ulcerans_Agy99 NAGAGIDIVQ-FNPKVVIYEISGPAGSTANINFWDADANTHQVNAAPLPW
Mb0696c|M.bovis_AF2122/97 TPKVFADDPEPFDPKVVEYEVSG-SGSYVNINYLDLDAKPQRIDGAALPW
Rv0677c|M.tuberculosis_H37Rv TPKVFADDPEPFDPKVVEYEVSG-SGSYVNINYLDLDAKPQRIDGAALPW
Mflv_3028|M.gilvum_PYR-GCK TP-VGSDTAEDFNPTVVTYEVFG-TGSFAVINYTDLDGLPQRTGQVSLPW
Mvan_1058|M.vanbaalenii_PYR-1 TP-VDADTAENFNPKVVTYEIFG-SGSTATINYADLDGKPQRTGEVSLPW
MLBr_02377|M.leprae_Br4923 YADSNLGSSKPFNPKQIVYEVFGPPGTVADISYFDANSDPQRIDGAQLPW
TH_2866|M.thermoresistible__bu YAGSTSVDTDKANPKRVRYEIFGAPGTVADINYIDAEGDPHQVNGATLPW
MAB_4311c|M.abscessus_ATCC_199 LGEGSG-ITANFNPKHITYEVVGS-GGTANLNYLDENGQPHLIENAPLPW
MSMEG_0462|M.smegmatis_MC2_155 VAEGTA-LTENFNPKHITYEVTGA-GGSANLNYLDENGQPHLVENAPLPW
MAV_4756|M.avium_104 SGEGAGDNTKPFHPKHITYEITGAPAGTVNANYLDENGQPHLVESAPLPW
.*. : **: * . . .: * :. .: . ***
MMAR_3743|M.marinum_M SFTISTTLPSVSANIMAQGDG--SQIGCRITVDGVVREEKSAAG-VNPQT
MUL_3687|M.ulcerans_Agy99 SFTISTTLPSVSANIMAQGDG--SQIGCRITVDGVVREEKSAAG-VNPQT
Mb0696c|M.bovis_AF2122/97 SLTLKTTAPSAAPNILAQGDG--TSITCRITVDGEVKDERTATG-VDALT
Rv0677c|M.tuberculosis_H37Rv SLTLKTTAPSAAPNILAQGDG--TSITCRITVDGEVKDERTATG-VDALT
Mflv_3028|M.gilvum_PYR-GCK TLTLETTIPSVRPDLLAQGNG--QFIGCRVTVDGEVEDERTAEG-VNAAT
Mvan_1058|M.vanbaalenii_PYR-1 SLTLETTLPSVMPNIMAQGDG--QSITCRVTVDDEVKDERTAEG-LNAAT
MLBr_02377|M.leprae_Br4923 SLLMTTTLAAVMGNLVAQGNT--DSIGCRIIVDGVVKAERVSNE-VNAYT
TH_2866|M.thermoresistible__bu SIEIVTNSPAMAGNVLAQGDS--DFIGCRIVSDGVVKDERTTQR-VSAYV
MAB_4311c|M.abscessus_ATCC_199 SFTIVTTLPSMSANIVAQGDRSVTDLGCRVTVDGQVRDDRSSADKVKPFI
MSMEG_0462|M.smegmatis_MC2_155 SFTIVTTLPSMSANIMAQGDRNVSGLRCRVTVDGEIRDDRASDDLVEPFI
MAV_4756|M.avium_104 SFTIVTTLPSMSANIVAQASEQVHALRCRVIVDDQVRDDRNTND-YQPFI
:: : *. .: :::**.. : **: *. :. :: : ..
MMAR_3743|M.marinum_M FCLVKSA
MUL_3687|M.ulcerans_Agy99 FCLVKSA
Mb0696c|M.bovis_AF2122/97 YCFVKSA
Rv0677c|M.tuberculosis_H37Rv YCFVKSA
Mflv_3028|M.gilvum_PYR-GCK YCLVKAA
Mvan_1058|M.vanbaalenii_PYR-1 YCLVKAA
MLBr_02377|M.leprae_Br4923 YCLVKSA
TH_2866|M.thermoresistible__bu YCFTKSA
MAB_4311c|M.abscessus_ATCC_199 YCLVKSV
MSMEG_0462|M.smegmatis_MC2_155 YCLVKSV
MAV_4756|M.avium_104 YCLVKSV
:*:.*:.