For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IDRSGRPPYTEDFPTTPHGYRLVHGPSAATRTRYAYGETFPDRDALAPNRLPRKRLRVAGMTFGAVAVAV VASATTAFAVVNHSGPAAPAGQVAAPVAGKPSPATGSTLGRPAATGAAGSVEEVSAKVLPSVVQLQIQSG RQQESGSGIVLSADGLILTNNHVVASAAPGAQPSAPVTEPDSPFGGQFGSLPPGIFPGEQQPGGYRDGRD GRDSDDYDGRGVRPSARTGDGVKATVTLSDGRTVPFTVVGADPDDDIAVVRAQGVNDLTPISIGSSKDLK VGQNVVAVGSPLGLQGTVTTGIISALNRPVATGDDQSGQRSVMSAIQTDAAINPGNSGGALVDMSGNLIG VNSAIASLSGGQDSQGGGQAGSIGLGFAIPVDQAKRIADELVSTGTVRHASLGVQLASSKDAPGAVVAGI VRGSAAATAGLPKGAVITKVDNQVIDGPDALVAAVRAKAPGDTMALTYQDPSGDARTVQVTLGQSEA*
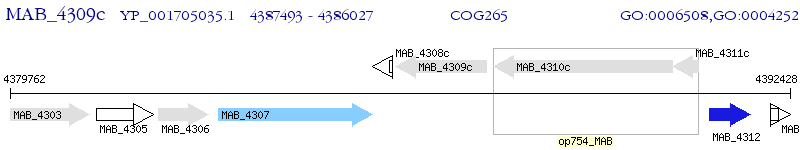
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4309c | - | - | 100% (488) | putative serine protease |
| M. abscessus ATCC 19977 | MAB_1078 | - | 1e-83 | 52.06% (340) | putative serine protease |
| M. abscessus ATCC 19977 | MAB_1364 | - | 2e-45 | 41.53% (248) | serine protease HtrA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1009 | - | 1e-118 | 52.77% (470) | serine protease |
| M. gilvum PYR-GCK | Mflv_1889 | - | 1e-106 | 49.89% (439) | peptidase S1 and S6, chymotrypsin/Hap |
| M. tuberculosis H37Rv | Rv0983 | pepD | 1e-117 | 52.55% (470) | serine protease PepD |
| M. leprae Br4923 | MLBr_00176 | - | 1e-115 | 53.95% (443) | putative secreted serine protease |
| M. marinum M | MMAR_4527 | pepD | 1e-114 | 53.95% (443) | serine protease PepD |
| M. avium 104 | MAV_1096 | - | 1e-117 | 55.71% (438) | protease |
| M. smegmatis MC2 155 | MSMEG_5486 | - | 1e-108 | 51.48% (439) | peptidase S1 and S6, chymotrypsin/Hap |
| M. thermoresistible (build 8) | TH_4473 | - | 1e-104 | 52.59% (405) | PUTATIVE peptidase S1 and S6, chymotrypsin/Hap |
| M. ulcerans Agy99 | MUL_4699 | pepD | 1e-114 | 53.72% (443) | serine protease PepD |
| M. vanbaalenii PYR-1 | Mvan_4842 | - | 1e-110 | 50.57% (441) | peptidase S1 and S6, chymotrypsin/Hap |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1009|M.bovis_AF2122/97 --------------------------------------------MAKLAR
Rv0983|M.tuberculosis_H37Rv --------------------------------------------MAKLAR
MMAR_4527|M.marinum_M --------------------------------------------------
MUL_4699|M.ulcerans_Agy99 --------------------------------------------------
MAV_1096|M.avium_104 --------------------------------------------------
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
Mflv_1889|M.gilvum_PYR-GCK MKMVEAGATGAEPDCCRKGNRTLRPFSVRLGTLSWQRSLFTRQDQFEGEH
Mvan_4842|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4309c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1009|M.bovis_AF2122/97 VVGLVQEEQPSDMTNH-PRYSPPPQQPGTP-------GYAQGQQQTYSQQ
Rv0983|M.tuberculosis_H37Rv VVGLVQEEQPSDMTNH-PRYSPPPQQPGTP-------GYAQGQQQTYSQQ
MMAR_4527|M.marinum_M ---MTNHPRYSPPPQQ-PGYRTAPNQPVPP-------AYAQGPQQNYNPQ
MUL_4699|M.ulcerans_Agy99 ---MTNHPGYSPPPQQ-PGYRTAPNQPVPP-------AYAQGPQQNYNPQ
MAV_1096|M.avium_104 ---MTNDPRYSPPPQQQPGYPPAPNQTAPVPSHAGSSGYAQG-QQPYQQQ
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
Mflv_1889|M.gilvum_PYR-GCK CSDMTNHPRYS--PTPQPGRQSGYTGQVPPGS--AGPAGSGSYEQQGTGS
Mvan_4842|M.vanbaalenii_PYR-1 ---MTNQPSYS--PTPQPGRQSGYPGQVPPGH--AGPR-TGSYEQQGSGG
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 ---MTNHPRYSQQPDPRTGATPGYPGGRPD-------------PYQQTSQ
MAB_4309c|M.abscessus_ATCC_199 ---MIDRSGRPPYTEDFPTTPHGYRLVHGP---------------SAATR
Mb1009|M.bovis_AF2122/97 FDWRYPPSPPPQPTQYR-----------------------QPYEALGGTR
Rv0983|M.tuberculosis_H37Rv FDWRYPPSPPPQPTQYR-----------------------QPYEALGGTR
MMAR_4527|M.marinum_M FDWRYHQSRPQPRPPYEGVSGTGPGLLPPATGPGPIPPDTGPGPIPGGTG
MUL_4699|M.ulcerans_Agy99 FDWRYHQSRPQPRPPYEGVSGTGPGLLPPATGPGPIPPDTGPGPIPGGTG
MAV_1096|M.avium_104 FDWRYR-SQPSQTTQFR--------------SPYEPFGNAGPGRVPGGTG
MLBr_00176|M.leprae_Br4923 --------------------------------------------------
Mflv_1889|M.gilvum_PYR-GCK WDWRYATEQQRQAFRGP-----------------YDSPYDPYRGAPMPGG
Mvan_4842|M.vanbaalenii_PYR-1 WDWRYATEQQRQAFR---------------------APYDPYAGAPMPSG
TH_4473|M.thermoresistible__bu --------------------------------------------------
MSMEG_5486|M.smegmatis_MC2_155 YDWRYAQQVQQ-------------------------APQSPQTHQTQPGF
MAB_4309c|M.abscessus_ATCC_199 TRYAYGETFPDRD-------------------------------------
Mb1009|M.bovis_AF2122/97 PGLIPGVIPTMTPPPGMVRQR-PRAGMLAIGAVTIAVVSAG--------I
Rv0983|M.tuberculosis_H37Rv PGLIPGVIPTMTPPPGMVRQR-PRAGMLAIGAVTIAVVSAG--------I
MMAR_4527|M.marinum_M PGPIPGMLPPMAPPPNVIRERRPRAGMLVMGAVVIAVVSAG--------I
MUL_4699|M.ulcerans_Agy99 PGPIPGMLPPMAPPPNVIRERRPRAGMLVMGAVVIAVVSAG--------I
MAV_1096|M.avium_104 TGPLPGMHPGMPGPP--EPQKRSRAGLLTVGAVAIAVVSAG--------I
MLBr_00176|M.leprae_Br4923 ------MIP-----PGRVADRRPRAGMLAVSALAIAVVSAG--------I
Mflv_1889|M.gilvum_PYR-GCK PGQFPPPGMP--RQPQPQPRKRS-GSALAAGAIALALVSGG--------I
Mvan_4842|M.vanbaalenii_PYR-1 PGHHPRPGMP--VTPPRQPRKGSRATALAAGAVAVALVSGG--------I
TH_4473|M.thermoresistible__bu ------------------------------------VVSAG--------I
MSMEG_5486|M.smegmatis_MC2_155 RPAYDPYRAPSTAQPVAIPAKKRSRAGLAAGALAVAVVSAG--------I
MAB_4309c|M.abscessus_ATCC_199 -----------ALAPNRLPRKRLRVAGMTFGAVAVAVVASATTAFAVVNH
:*:..
Mb1009|M.bovis_AF2122/97 GGAAASLVGFNRAPAGPSGGPVAASAAPSIPAANMPPGSVEQVAAKVVPS
Rv0983|M.tuberculosis_H37Rv GGAAASLVGFNRAPAGPSGGPVAASAAPSIPAANMPPGSVEQVAAKVVPS
MMAR_4527|M.marinum_M GGAAASIIELNRSPATSSGREMVASGAPSVPAANMPPGSVEQVAAKVLPS
MUL_4699|M.ulcerans_Agy99 GGAAASIIELNRSPATSSGREMVASGAPSVPAANMPPGSVEQVAAKVLPS
MAV_1096|M.avium_104 GGAAATAVEMG-THATGSGGGRIAGAAPSVPAANMPPGSVEQVAAKVVPS
MLBr_00176|M.leprae_Br4923 GGVAATVVALGTRVAGGNGAGPVTGPAASVPAANMPSGSVEQVAVKVVPS
Mflv_1889|M.gilvum_PYR-GCK GGGVAMLAHPDHSLGGIS----ASGAAPSMPAASVPGGSVEAVAAKVVPS
Mvan_4842|M.vanbaalenii_PYR-1 GGGVAMLVHPDHGVPGIS----ASGAAPGMPAASVPAGSVEAVAAAVVPS
TH_4473|M.thermoresistible__bu GGGVATLAQPERPADYSS----LSDAASAVPAASIPPGTVEQVAAKVVPS
MSMEG_5486|M.smegmatis_MC2_155 GGGVATLVHNESPQLGSP----AIGAAPTVPAAALPAGSVEQVAAKVVPS
MAB_4309c|M.abscessus_ATCC_199 SGPAAPAGQVAAPVAGKPS--PATGSTLGRPAATGAAGSVEEVSAKVLPS
.* .* . : *** . *:** *:. *:**
Mb1009|M.bovis_AF2122/97 VVMLETDLGRQSEEGSGIILSAEGLILTNNHVIAAAAKPP----------
Rv0983|M.tuberculosis_H37Rv VVMLETDLGRQSEEGSGIILSAEGLILTNNHVIAAAAKPP----------
MMAR_4527|M.marinum_M VVMLETDVGRQSEEGSGIILSADGLVMTNNHVVAAAAKPP----------
MUL_4699|M.ulcerans_Agy99 VVMLETDVGRQSEEGSGIILSADGLVMTNNHVVAAAAKPP----------
MAV_1096|M.avium_104 VVMLETDLGRQSEEGSGIILSTDGLILTNNHVVAAAAGPPKGPAAPPVVP
MLBr_00176|M.leprae_Br4923 VVMLETDLGRQSEEGSGVILSADGLILTNNHVVAVAAKPGGGP-------
Mflv_1889|M.gilvum_PYR-GCK VVKLEVNQGRSTEEGSGVILSSDGLILTNNHVVAAAADNG----------
Mvan_4842|M.vanbaalenii_PYR-1 VVKLEVSQGRASEEGSGVILSTDGLILTNNHVVATAAG------------
TH_4473|M.thermoresistible__bu VVKLETELGRATEEGSGIILSSDGLILTNHHVVSAAVREP----------
MSMEG_5486|M.smegmatis_MC2_155 VVKLETDLGRASEEGSGIILTADGLILTNNHVVAAAKG------------
MAB_4309c|M.abscessus_ATCC_199 VVQLQIQSGRQQESGSGIVLSADGLILTNNHVVASAAPGAQPSAPVTEPD
** *: . ** *.***::*:::**::**:**:: *
Mb1009|M.bovis_AF2122/97 ---------------------------------------------LGSPP
Rv0983|M.tuberculosis_H37Rv ---------------------------------------------LGSPP
MMAR_4527|M.marinum_M ---------------------------------------------AGAPE
MUL_4699|M.ulcerans_Agy99 ---------------------------------------------AGAPE
MAV_1096|M.avium_104 LPPG------------------------------------GPGGGPGAGT
MLBr_00176|M.leprae_Br4923 ---------------------------------------------GGGLS
Mflv_1889|M.gilvum_PYR-GCK -----------------------------------------APGAPPGA-
Mvan_4842|M.vanbaalenii_PYR-1 -----------------------------------------AAGEPGGP-
TH_4473|M.thermoresistible__bu -----------------------------------------GRTDAQTPN
MSMEG_5486|M.smegmatis_MC2_155 --------------------------------------------DGPGPA
MAB_4309c|M.abscessus_ATCC_199 SPFGGQFGSLPPGIFPGEQQPGGYRDGRDGRDSDDYDGRGVRPSARTGDG
Mb1009|M.bovis_AF2122/97 PKTTVTFSDGRTAPFTVVGADPTSDIAVVRVQGVSGLTPISLGSSSDLRV
Rv0983|M.tuberculosis_H37Rv PKTTVTFSDGRTAPFTVVGADPTSDIAVVRVQGVSGLTPISLGSSSDLRV
MMAR_4527|M.marinum_M PKTTVTLYDGRTASFTVVGADPTSDIAVVRVSGVSGLTPIALGSSADLRV
MUL_4699|M.ulcerans_Agy99 PKTTVTLYDGRTASFTVVGADPTSDIAVVRVSGVSGLTPIALGSSADLRV
MAV_1096|M.avium_104 PKTTVTFSDGRTAPFTVVGADPTSDIAVIRVQGMSGLTPISLGSSSDLRV
MLBr_00176|M.leprae_Br4923 PKTTVTFFDGRTASFTVVGADPTSDIAVVRVQSISGLTPITMGSSADLRV
Mflv_1889|M.gilvum_PYR-GCK AQTKVTFADGKSSTFEVIGTDPGSDIAVVRAKNVTDLTPIAVGSSADLRV
Mvan_4842|M.vanbaalenii_PYR-1 AKTKVTFADGKTAPFTVIGADPSSDIAVVRAQNVSGLTPITVGSSADLRV
TH_4473|M.thermoresistible__bu VRTKVTFADGRTAPFTVVGSDPSSDIAVVRATNVTGLTPVTLGSSADLRV
MSMEG_5486|M.smegmatis_MC2_155 AQTKVTFADGTTRPFTVVGTDPSSDIAVVRAQDVSGLTPITLGSSANLRV
MAB_4309c|M.abscessus_ATCC_199 VKATVTLSDGRTVPFTVVGADPDDDIAVVRAQGVNDLTPISIGSSKDLKV
::.**: ** : .* *:*:** .****:*. .:..***:::*** :*:*
Mb1009|M.bovis_AF2122/97 GQPVLAIGSPLGLEGTVTTGIVSALNRPVSTTGEAGNQNTVLDAIQTDAA
Rv0983|M.tuberculosis_H37Rv GQPVLAIGSPLGLEGTVTTGIVSALNRPVSTTGEAGNQNTVLDAIQTDAA
MMAR_4527|M.marinum_M GQPVLAIGSPLGLAGTVTTGIVSALNRPVSTTGEAGNQNTVLDAIQTDAA
MUL_4699|M.ulcerans_Agy99 GQPLLAIGSPLGLAGTVTTGIVSALNRPVSTTGESGNQNTVLDAIQTDAA
MAV_1096|M.avium_104 GQPVVAIGSPLGLSGTVTTGIVSALNRPVSTTGESGNQNTVLDAIQTDAA
MLBr_00176|M.leprae_Br4923 GQPVVAVGSPLGLAGTVTSGIVSALNRPVSTTGESGNQNTVLDAIQTDAA
Mflv_1889|M.gilvum_PYR-GCK GQDVVAIGSPLGLEGTVTTGIISALNRPVAASGDARNQNTVLDAIQTDAA
Mvan_4842|M.vanbaalenii_PYR-1 GQDVVAIGSPLGLEGTVTTGIISALNRPVAAGGDARNQNTVLDAIQTDAA
TH_4473|M.thermoresistible__bu GQDVVAIGSPLGLEGTVTTGIVSALNRPVAAGGDARNQNTVLDAIQTDAA
MSMEG_5486|M.smegmatis_MC2_155 GQDVVAIGSPLGLEGTVTTGIVSALNRPVAAGGDTRNQNTVLDAIQTDAA
MAB_4309c|M.abscessus_ATCC_199 GQNVVAVGSPLGLQGTVTTGIISALNRPVATGDDQSGQRSVMSAIQTDAA
** ::*:****** ****:**:*******:: .: .*.:*:.*******
Mb1009|M.bovis_AF2122/97 INPGNSGGALVNMNAQLVGVNSAIATLGADS---ADAQSGSIGLGFAIPV
Rv0983|M.tuberculosis_H37Rv INPGNSGGALVNMNAQLVGVNSAIATLGADS---ADAQSGSIGLGFAIPV
MMAR_4527|M.marinum_M INPGNSGGALVNMNAQLVGINSAIATLGADS---GDAQSGSIGLGFAIPV
MUL_4699|M.ulcerans_Agy99 INPGNSGGALVNMNAQLVGINSAIATLGADS---GDAQSGSIGLGFAIPV
MAV_1096|M.avium_104 INPGNSGGALVNMNGQLVGVNSAIATLGADS---PDAQSGSIGLGFAIPV
MLBr_00176|M.leprae_Br4923 INPGNSGGALVNMGGQLVGVNSAIATLGADS---GDAQSGSIGLGFAIPV
Mflv_1889|M.gilvum_PYR-GCK INPGNSGGALVNMNGELVGINSAIATMGADA---GGPQGGSIGLGFAIPV
Mvan_4842|M.vanbaalenii_PYR-1 INPGNSGGALVNMNGELVGINSAIATMGADA---GAQQGGSIGLGFAIPV
TH_4473|M.thermoresistible__bu INPGNSGGALVNMNGELVGVNSAIATLGGGV---AQTQSGSIGLGFAIPV
MSMEG_5486|M.smegmatis_MC2_155 INPGNSGGALVNMNGELVGINSAIATMGGDS---PNAQSGSIGLGFAIPV
MAB_4309c|M.abscessus_ATCC_199 INPGNSGGALVDMSGNLIGVNSAIASLSGGQDSQGGGQAGSIGLGFAIPV
***********:*..:*:*:*****::... *.***********
Mb1009|M.bovis_AF2122/97 DQAKRIADELISTGKASHASLGVQVTNDKDTPGAKIVEVVAGGAAANAGV
Rv0983|M.tuberculosis_H37Rv DQAKRIADELISTGKASHASLGVQVTNDKDTLGAKIVEVVAGGAAANAGV
MMAR_4527|M.marinum_M DQAKRIADELISTGKASHASLGVQVTTDKGVPGAKVVEVVQGGAAATAGL
MUL_4699|M.ulcerans_Agy99 DQAKRIADELISTGKASHASLGVQVTTDKGVPGAKVVEVVQGGAAATAGL
MAV_1096|M.avium_104 DQAKRIADELIATGKASHASLGVQVTNDKGTPGAKVVDVVSGGAAASAGL
MLBr_00176|M.leprae_Br4923 DQAKRIADELISTGKATHASLGVQVATDKGTPGAKVMDVVAGGAAANAAV
Mflv_1889|M.gilvum_PYR-GCK DQAKRIADEIIQTGSASRASLGVQVGNEAGVDGAKIVEVTAGGAASAAGL
Mvan_4842|M.vanbaalenii_PYR-1 DQAKRIADEIIQTGSASRASLGVQVGNEAGVDGAKIVEVTAGGAASAAGL
TH_4473|M.thermoresistible__bu DQAKRIADELIKHGRATHASLGVQVSNEAGTDGAMIVEVTEGGAAAAAGL
MSMEG_5486|M.smegmatis_MC2_155 DQAKRIADELIQNGVASHASLGVQVANDRSSDGAKIVEVNDGGAAAAAGL
MAB_4309c|M.abscessus_ATCC_199 DQAKRIADELVSTGTVRHASLGVQLASSKDAPGAVVAGIVRGSAAATAGL
*********:: * . :******: .. . ** : : *.**: *.:
Mb1009|M.bovis_AF2122/97 PKGVVVTKVDDRPINSADALVAAVRSKAPGATVALTFQDPSGG-SRTVQV
Rv0983|M.tuberculosis_H37Rv PKGVVVTKVDDRPINSADALVAAVRSKAPGATVALTFQDPSGG-SRTVQV
MMAR_4527|M.marinum_M PKGVVVTKVDDRAITSADALVAAVRSKAPGDKVSLTFQDPDGS-TRTVQV
MUL_4699|M.ulcerans_Agy99 PKGVVVTKVDDRAITSADALVAAVRSKAPGDKVSLTFQDPDGS-TRTVQV
MAV_1096|M.avium_104 PKNVVVTKVDDRPINSADALVAAVRSKAPGDKITLTFSDPAGGGSRTLPV
MLBr_00176|M.leprae_Br4923 PKGVVLTKVDDRLISSADALVAAVRSKAPGDKVSLTYQDQSGS-SRTVQV
Mflv_1889|M.gilvum_PYR-GCK PSGVIVTKLDDRAISSADALVAAVRSKAPGDKVTLTYQDASGK-PRTVDV
Mvan_4842|M.vanbaalenii_PYR-1 PSGVIVTKVDDRLINSADALVAAVRSKAPGDKVTLTYLDPAGK-SQSLDV
TH_4473|M.thermoresistible__bu PAGVVVTKLDDRLINSADALVAAVRSKAPGDTVTLTYVEGSGK-PRTVDV
MSMEG_5486|M.smegmatis_MC2_155 PSGVVVTKVDDRVIGSADALVAAVRSKAPGEKVTLTYLDQAGK-PQTVDV
MAB_4309c|M.abscessus_ATCC_199 PKGAVITKVDNQVIDGPDALVAAVRAKAPGDTMALTYQDPSGD-ARTVQV
* ..::**:*:: * ..********:**** .::**: : * .::: *
Mb1009|M.bovis_AF2122/97 TLGKAEQ
Rv0983|M.tuberculosis_H37Rv TLGKAEQ
MMAR_4527|M.marinum_M TLGKAEQ
MUL_4699|M.ulcerans_Agy99 TLGKAEQ
MAV_1096|M.avium_104 TLGKADQ
MLBr_00176|M.leprae_Br4923 TLGKAEQ
Mflv_1889|M.gilvum_PYR-GCK TLGKAKQ
Mvan_4842|M.vanbaalenii_PYR-1 TLGKAGQ
TH_4473|M.thermoresistible__bu TLGKMQR
MSMEG_5486|M.smegmatis_MC2_155 TLGKAEQ
MAB_4309c|M.abscessus_ATCC_199 TLGQSEA
***: