For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRYAMQGTARRFGALAFVAALLAGCSTPENSTPLPQLTVPRPTPAEMTEQEPNRAVLDKIDTSCDPTAS LRPTSDRAANDEAVAAIRKRGRLVVGLDIGSNLFSFRDPITGELTGFDVDIAGEVSRDIFGTPERVEYRI LSSADRVAALKNSTVDIVVKTMTITCARRKEVNFSSVYLMNYQRILTSRDSGITNAQDLSGRRVCAARGT TSLNRLSQITPPPVIMSVVTWADCLVALQQRQVDAVSTDDSILAGLMSQDPYLHIIGPNMSDEPYGIGIK LENTDLVRYVNRTLERIRRDGTWNALYRKWLSVLGPAPAPPSAKYRD
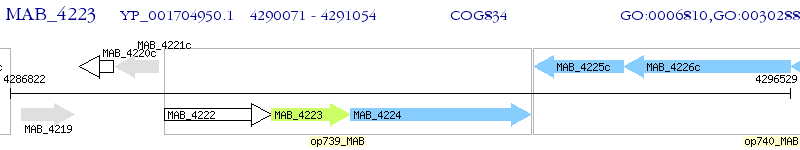
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4223 | - | - | 100% (327) | glutamine-binding protein GlnH |
| M. abscessus ATCC 19977 | MAB_3051 | - | 3e-18 | 25.99% (227) | putative glutamate ABC transporter, periplasmic protein |
| M. abscessus ATCC 19977 | MAB_0277c | - | 3e-14 | 29.15% (223) | amino acid ABC transporter permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0419c | glnH | 1e-119 | 65.85% (328) | glutamine-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_0201 | - | 1e-124 | 67.49% (323) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv0411c | glnH | 1e-119 | 65.85% (328) | glutamine-binding lipoprotein |
| M. leprae Br4923 | MLBr_00303 | glnH | 1e-116 | 63.53% (329) | putative glutamine-binding protein |
| M. marinum M | MMAR_0714 | glnH | 1e-118 | 66.26% (329) | glutamine-binding lipoprotein GlnH |
| M. avium 104 | MAV_4750 | - | 1e-117 | 67.51% (317) | extracellular solute-binding protein, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_0787 | - | 1e-123 | 71.52% (309) | extracellular solute-binding protein, family protein 3 |
| M. thermoresistible (build 8) | TH_2308 | glnH | 1e-119 | 65.72% (318) | PROBABLE GLUTAMINE-BINDING LIPOPROTEIN GLNH (GLNBP) |
| M. ulcerans Agy99 | MUL_2809 | glnH | 1e-118 | 66.26% (329) | glutamine-binding lipoprotein GlnH |
| M. vanbaalenii PYR-1 | Mvan_0704 | - | 1e-124 | 68.35% (316) | extracellular solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0201|M.gilvum_PYR-GCK ----------MMKRALRRV---GAALAAAVMLSGCAQALPVVPTPTVTLA
Mvan_0704|M.vanbaalenii_PYR-1 -----MSACAKKKQPLMKVSAWGALLAGVLVLGGCAQTSPVVPTPSVTLA
MSMEG_0787|M.smegmatis_MC2_155 -----------MKRLLMLL----VSTLTAMSLVGCSQAAPALNLPSLTLA
TH_2308|M.thermoresistible__bu ----MDIEVRTMGTPVKRL--VAVLGVLGMLLTGCGESNSVAPAPEVVLN
Mb0419c|M.bovis_AF2122/97 ---------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLP
Rv0411c|M.tuberculosis_H37Rv ---------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLP
MMAR_0714|M.marinum_M ---------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLP
MUL_2809|M.ulcerans_Agy99 ---------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLP
MLBr_00303|M.leprae_Br4923 --------MMTRQFKIQRACS---VLATAMMLAACGHAELLTVASMPTLP
MAV_4750|M.avium_104 MAEAERVPMTAGLPLLRRACT---ALAAVLVAAGCGHTESLRVASVPTLP
MAB_4223|M.abscessus_ATCC_1997 ---------MTRYAMQGTARRFGALAFVAALLAGCSTPENSTPLPQLTVP
.*. . . .:
Mflv_0201|M.gilvum_PYR-GCK PPTPAGLEELPPVPARPATPADTDCDPRASLRPFATQEEADAAVANIKAR
Mvan_0704|M.vanbaalenii_PYR-1 PPTPAGLEEMPPEPARAPTAADDDCDPLASLRPFDNKEDADKAVANIKAR
MSMEG_0787|M.smegmatis_MC2_155 PPTPAGMEEIPPQPVRLPEIDNDDCNRTASLRPFSNKEEAEAAVANIRAR
TH_2308|M.thermoresistible__bu PPSPSGMEEMPPQPAQMPDPKAEDCDRTASLRPFPTRAEADEAVAHIRER
Mb0419c|M.bovis_AF2122/97 LPTPVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRAR
Rv0411c|M.tuberculosis_H37Rv LPTPVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRAR
MMAR_0714|M.marinum_M LPTPVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRAR
MUL_2809|M.ulcerans_Agy99 LPTPVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRAR
MLBr_00303|M.leprae_Br4923 PPTPVGMEQVPPELLLPQDNTEEDCDPTASLRPFPTKAEADAAVAEIRAR
MAV_4750|M.avium_104 PPTPVGMEQLPPQPPLPPDGPDQNCDLTASLRPFPTKAEADAAVADIRAR
MAB_4223|M.abscessus_ATCC_1997 RPTPAEMTEQEPNRAVL-DKIDTSCDPTASLRPTSDRAANDEAVAAIRKR
*:* : * .*: ***** : : *** *: *
Mflv_0201|M.gilvum_PYR-GCK GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEIARDIFGTPSQVEYRI
Mvan_0704|M.vanbaalenii_PYR-1 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEIARDIFGTPSQVEYRI
MSMEG_0787|M.smegmatis_MC2_155 GRLIVGLDIGSNLFSFRDPITGQITGFDVDIAGEVSRDIFGTPSKVEYRI
TH_2308|M.thermoresistible__bu GRLIVGLDIGSNLFSFRDPITGQIAGFDVDIAGEIARDIFGSPAQVEYRI
Mb0419c|M.bovis_AF2122/97 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRI
Rv0411c|M.tuberculosis_H37Rv GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRI
MMAR_0714|M.marinum_M GRLIVGLDIGSNLFSFRDPITGEITGFDVDIASEVARDIFGGPTHVEYRI
MUL_2809|M.ulcerans_Agy99 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIASEVARDIFGGPTHVEYRI
MLBr_00303|M.leprae_Br4923 GRLIVGLEIGSNLFSFRDPITGEITGFDIDIAGEVARDIFGTPSHMEYRI
MAV_4750|M.avium_104 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEIARDIFGAPSHVEYRI
MAB_4223|M.abscessus_ATCC_1997 GRLVVGLDIGSNLFSFRDPITGELTGFDVDIAGEVSRDIFGTPERVEYRI
***:***:**************:::***:***.*::***** * ::****
Mflv_0201|M.gilvum_PYR-GCK LSSADRVDALRDNQVDVVVKTMTITCERKKLVNFSTAYLIANQRILAPRD
Mvan_0704|M.vanbaalenii_PYR-1 LSSADRVEALQKNQVDVVVKTMTITCERKKLVNFSTAYLSANQRILAPRD
MSMEG_0787|M.smegmatis_MC2_155 LSSADRITALRNNQVDIVVKTMTITCERKKLVNFSTVYLMANQRILASRD
TH_2308|M.thermoresistible__bu LSSADRIEALQNNEVDVVVKTMTITCERKEKIHFSTAYLNANQRILAPRS
Mb0419c|M.bovis_AF2122/97 LSAAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRILAPRD
Rv0411c|M.tuberculosis_H37Rv LSAAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRILAPRD
MMAR_0714|M.marinum_M LSAAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRILTSRD
MUL_2809|M.ulcerans_Agy99 LSAAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRILTSRD
MLBr_00303|M.leprae_Br4923 LSTAARITALQKSEVDIIVKTMTITCKRRKLVNFSTVYLDAKQRILAPRD
MAV_4750|M.avium_104 LSSDERVTALQRGEVDVVVKTMTITCDRRKQVNFSTVYLDANQRILAPRD
MAB_4223|M.abscessus_ATCC_1997 LSSADRVAALKNSTVDIVVKTMTITCARRKEVNFSSVYLMNYQRILTSRD
**: *: **: . **::****:*** *:: ::**:.** ****:.*.
Mflv_0201|M.gilvum_PYR-GCK SNIRQSSDLSGKRVCVAKGTTSLERIQQISPPPLIVGVVTWADCLVALQQ
Mvan_0704|M.vanbaalenii_PYR-1 SNIRQSSDLSGKRVCVAKGTTSLERIQQITPPPIIVGVVTWADCLVALQQ
MSMEG_0787|M.smegmatis_MC2_155 SNISQASDLSGRRVCVVDGTTSLQRIQQISPSPIIVSVVTWADCLVALQQ
TH_2308|M.thermoresistible__bu SPITRAADLSGRRVCVVKGTTSLTRVQQISPPPIIVAVETWADCLVAIQQ
Mb0419c|M.bovis_AF2122/97 SPITKVSDLSGKRVCVARGTTSLRRIREIAPPPVIVSVVNWADCLVALQQ
Rv0411c|M.tuberculosis_H37Rv SPITKVSDLSGKRVCVARGTTSLRRIREIAPPPVIVSVVNWADCLVALQQ
MMAR_0714|M.marinum_M SPIFKVSDLSGKRVCVARGTTSLRRIREIAPPPVIVSVVNWADCLVALQQ
MUL_2809|M.ulcerans_Agy99 SPIFKVSDLSGKRVCVARGTTSLRRIREIAPPPVIVSVVNWADCLVALQQ
MLBr_00303|M.leprae_Br4923 SPITKPSDLSNKRVCMTRGTTSLHQVSEVVPPPIIVSAENWADCLVAMQQ
MAV_4750|M.avium_104 SPITKVSDLSGKRVCVAKGTTSLHRIRQIDPPPIVVSVVNWADCLVAMQQ
MAB_4223|M.abscessus_ATCC_1997 SGITNAQDLSGRRVCAARGTTSLNRLSQITPPPVIMSVVTWADCLVALQQ
* * . ***.:*** . ***** :: :: *.*:::.. .*******:**
Mflv_0201|M.gilvum_PYR-GCK RQVDAVSTDDAILAGLVSQDPYLHIVGPSMNEEPYGIGVNLQNTGLVRYV
Mvan_0704|M.vanbaalenii_PYR-1 RQVDAVSTDDSILAGLVSQDPYLHIVGPSMNEEPYGIGVNLENTGLVRFV
MSMEG_0787|M.smegmatis_MC2_155 RQADAVSTDDSILAGLVAQDPYLHIVGPSLSQEPYGIGVNLENTGLVRVV
TH_2308|M.thermoresistible__bu NQVDAVSTDDSILAGLMAQDPYLHIVGPSMSQEPYGIGINKNNPGLVRFV
Mb0419c|M.bovis_AF2122/97 REIDAVSTDDTILAGLVEEDPYLHIVGPDMADQPYGVGINLDNTGLVRFV
Rv0411c|M.tuberculosis_H37Rv REIDAVSTDDTILAGLVEEDPYLHIVGPDMADQPYGVGINLDNTGLVRFV
MMAR_0714|M.marinum_M REIDAVSTDDTILAGLVEEDPYLHIVGPNMANQPYGIGINLDNTGLVRFV
MUL_2809|M.ulcerans_Agy99 REIDAVSTDDTILAGLVEEDPYLHIVGPNMANQPYGIGINLDNTGLVRFV
MLBr_00303|M.leprae_Br4923 REIDAISTDDAILAGLVEEDPYLHIVGPNMATQPYGIGINLNNTTLVRFV
MAV_4750|M.avium_104 REIDAVSTDDSILAGLVEEDPYLHIVGPNMATQPYGIGINLNNTGLVRFV
MAB_4223|M.abscessus_ATCC_1997 RQVDAVSTDDSILAGLMSQDPYLHIIGPNMSDEPYGIGIKLENTDLVRYV
.: **:****:*****: :******:**.: :***:*:: :*. *** *
Mflv_0201|M.gilvum_PYR-GCK NGTLQRIRRDGTWNTLYRKWLTVLGPAPAPPVARYTD
Mvan_0704|M.vanbaalenii_PYR-1 NGTLQRIRRDGTWNTLYRKWLTVLGPAPAPPAARYSD
MSMEG_0787|M.smegmatis_MC2_155 NGTLERIRRDGTWYTLYRKWLTVLGPAPAPPVARYVD
TH_2308|M.thermoresistible__bu NGTLERIRRDGTWNTLYRKWLTVLGPAPAPPVARYVD
Mb0419c|M.bovis_AF2122/97 NGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD
Rv0411c|M.tuberculosis_H37Rv NGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD
MMAR_0714|M.marinum_M NGTLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD
MUL_2809|M.ulcerans_Agy99 NGTLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD
MLBr_00303|M.leprae_Br4923 NGTLERIRRDGTWITLYRKWLTVLGPAPAPPTPRYVD
MAV_4750|M.avium_104 NGTLERIRRDGTWNTLYRKWLTVLGPAPAPPTPRYLD
MAB_4223|M.abscessus_ATCC_1997 NRTLERIRRDGTWNALYRKWLSVLGPAPAPPSAKYRD
* **:***.**** :******:********* .:* *