For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLPAVSPTHLIGFAVAAYALIVIPGPSVLFAVGRSLSLGRRSGLVTVLGNTTGTVVPLVLTTAGLGAVLA ASTWALTVVKLVGAAYLIYLGAQAIRNRKSLVTALDAAAPAVGSQRVFRQGFVVGMTNPKTALFFAAVLP QFADPAVGSVPMQMAVFGAIFVLIALVSDSVWAVLASTARNWFARSPKRLEAVGATGGWMIVGLGAGVAV SSP
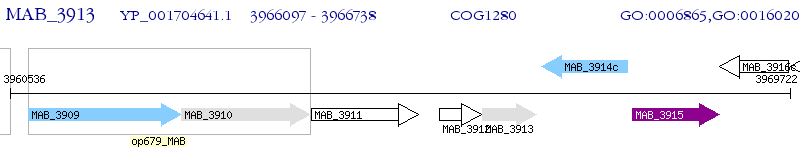
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3913 | - | - | 100% (213) | putative translocator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2008 | - | 5e-05 | 26.99% (163) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3615 | - | 4e-18 | 31.87% (182) | lysine exporter protein LysE/YggA |
| M. tuberculosis H37Rv | Rv1986 | - | 5e-05 | 26.99% (163) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3203 | - | 4e-21 | 34.30% (172) | transporter, LysE family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0172 | - | 4e-22 | 31.03% (203) | lysine exporter protein LysE/YggA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3615|M.gilvum_PYR-GCK --------MWLAFFGAAVVIAVSPGAGAIQSMATGLTHGVTRGYWSIVGL
MSMEG_3203|M.smegmatis_MC2_155 ----MEWHVWLAFFGAAIAISVSPGAGAIQSMATGLTHGVRRGYWSILGL
Mvan_0172|M.vanbaalenii_PYR-1 --------MTLAFLLTSLVIVATPGTGALYTIATGLAHGTRASVVAAVGC
Mb2008|M.bovis_AF2122/97 -----MNSPLVVGFLACFTLIAAIGAQNAFVLRQGIQR--EHVLPVVALC
Rv1986|M.tuberculosis_H37Rv -----MNSPLVVGFLACFTLIAAIGAQNAFVLRQGIQR--EHVLPVVALC
MAB_3913|M.abscessus_ATCC_1997 MLPAVSPTHLIGFAVAAYALIVIPGPSVLFAVGRSLSLGRRSGLVTVLGN
: :. .: . *. : .:
Mflv_3615|M.gilvum_PYR-GCK EAGLMLQLTLVAVGLGAAVANSVLAFTVIKWLGVVYLAYLAWRQWRSVAV
MSMEG_3203|M.smegmatis_MC2_155 EIGLMLQLALVAIGLGAAVAGSIVAFNIIKWIGVAYLIYLAVRQWRTAAI
Mvan_0172|M.vanbaalenii_PYR-1 TIGIVPAMLAAVTGLAAILHSSAIAFQTIKWLGVGYLLYLAWITWR-DKT
Mb2008|M.bovis_AF2122/97 TVSDIVLIAAGIAGFGALIGAHPRALNVVKFGGAAFLIGYGLLAARRAWR
Rv1986|M.tuberculosis_H37Rv TVSDIVLIAAGIAGFGALIGAHPRALNVVKFGGAAFLIGYGLLAARRAWR
MAB_3913|M.abscessus_ATCC_1997 TTGTVVPLVLTTAGLGAVLAASTWALTVVKLVGAAYLIYLGAQAIRNRKS
. : : *:.* : *: :* *. :* . *
Mflv_3615|M.gilvum_PYR-GCK DLHSQVDRGMDGGRVSLMVRGFLVNATNPKGLLFFLAVMPQFVNPAAPLL
MSMEG_3203|M.smegmatis_MC2_155 DLREQLGKATEAGRTGLVVRGFLVNATNPKGLVFFLAVLPQFVVPTAPLL
Mvan_0172|M.vanbaalenii_PYR-1 ELVADAVSSERPGSWRITGTAVAVNVLNPKLTIFFFAFLPQFVDPARPPV
Mb2008|M.bovis_AF2122/97 PVALIPSGATPVRLAEVLVTCAAFTFLNPHVYLDTVVLLGALANEHS---
Rv1986|M.tuberculosis_H37Rv PVALIPSGATPVRLAEVLVTCAAFTFLNPHVYLDTVVLLGALANEHS---
MAB_3913|M.abscessus_ATCC_1997 LVTALDAAAPAVGSQRVFRQGFVVGMTNPKTALFFAAVLPQFADPAVGSV
: . : . **: : ..: :.
Mflv_3615|M.gilvum_PYR-GCK -AQYLAIGVTMVAVDVVVMGFYTALSVRLLTWLRT-PRQQMTLNRVFSGL
MSMEG_3203|M.smegmatis_MC2_155 -PQYLAIGATMIAVDLVVMGLYTGLAVRLLTWLHT-PRQQTILNRVFSGL
Mvan_0172|M.vanbaalenii_PYR-1 -LQMLYLSAIFMAMTLVVFALYGAFAAGVRTHVISRPRVVTWMRRTFAAT
Mb2008|M.bovis_AF2122/97 -DQRWLFGLGAVTASAVWFATLG-FGAGRLRGLFTNPGSWRILDGLIAVM
Rv1986|M.tuberculosis_H37Rv -DQRWLFGLGAVTASAVWFATLG-FGAGRLRGLFTNPGSWRILDGLIAVM
MAB_3913|M.abscessus_ATCC_1997 PMQMAVFGAIFVLIALVSDSVWAVLASTARNWFARSPKRLEAVGATGGWM
* :. : * . :. . * : .
Mflv_3615|M.gilvum_PYR-GCK FATAAVVLALVRRGPAAA
MSMEG_3203|M.smegmatis_MC2_155 FAAAAVVLSLVRRAATV-
Mvan_0172|M.vanbaalenii_PYR-1 YVALAARMAVTTR-----
Mb2008|M.bovis_AF2122/97 MVALGISLTVT-------
Rv1986|M.tuberculosis_H37Rv MVALGISLTVT-------
MAB_3913|M.abscessus_ATCC_1997 IVGLGAGVAVSSP-----
. . :::