For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TATAQPAASTSPYLSGIWEPVQQEMDSVDLEVTGTLPAHLDGRYLRNGPNPAAEVDPATYHLFSGDAMVH GLSLREGKAQWYRNRWVRTPSVSKTLGETRPAAISANAGMGVIGPNTNVLGHAGRTLALVEGGIANYELT EDLDTIGTCDFDGTLPGGYTAHPHTDPDTGEMHAVSYSFARGSTVQYSVIDAHGRARRTVDIPVHGAPMM HDFTLTEKYVVFYDLPVTLDMSMISQSVPVPRLLRKPAQIVMNSLIGKVKIPGPIAAKASQYSGKSPSIP YSWNPKYPARIGVMPREGDDASRHAPVRWFEIDPCYVFHPLNGYSEMRNGAEVIVIDLVRYDSMFSQDVL GPSDAKGQLDRWTIDLAKGTVHMERKDDRHLEFPRINETFTGKKHRYGYLPDVENFFGSGEAAGASIVKY DYETGSTLSSRLDPAIVLGEMSFVPNPTCAAEDDGVLIGFAVDRRSDEGQLLLLDATSLDLMATVHLPQR VPAGFHGNWAPR*
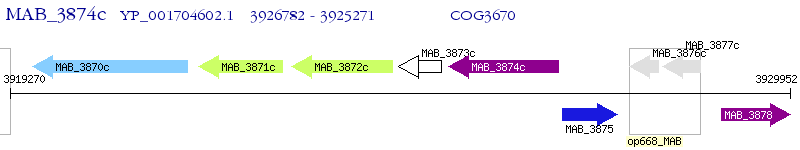
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3874c | - | - | 100% (503) | putative dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0673 | - | 0.0 | 61.83% (503) | dioxygenase |
| M. gilvum PYR-GCK | Mflv_2111 | - | 1e-163 | 56.24% (489) | carotenoid oxygenase |
| M. tuberculosis H37Rv | Rv0654 | - | 0.0 | 61.83% (503) | dioxygenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0993 | - | 0.0 | 62.00% (500) | dioxygenase |
| M. avium 104 | MAV_4505 | - | 1e-177 | 59.57% (507) | Sim14 protein |
| M. smegmatis MC2 155 | MSMEG_6224 | - | 8e-77 | 37.50% (488) | retinal pigment epithelial membrane protein |
| M. thermoresistible (build 8) | TH_3959 | - | 1e-164 | 55.62% (489) | PUTATIVE Carotenoid oxygenase |
| M. ulcerans Agy99 | MUL_0744 | - | 0.0 | 61.80% (500) | dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_4595 | - | 1e-160 | 54.81% (489) | carotenoid oxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2111|M.gilvum_PYR-GCK -----------MANRYLEGAFAPISEEHTLTDLDVIGTIPDHLDGRYLRN
TH_3959|M.thermoresistible__bu -----------VSNRYLTGAFAPLTEEYTLTDLEVTGTIPDYLDGRYLRN
Mvan_4595|M.vanbaalenii_PYR-1 -----------MGNQYLEGAFAPVANEYTLTGLDVVGDIPAYLDGRYLRN
MMAR_0993|M.marinum_M ---MTTMRSAQPRNPYLEGFLAPVRAEVTATDLEVTGRIPDYLDGRYLRN
MUL_0744|M.ulcerans_Agy99 ---MTTMRSAQPRNPYLEGFLAPVRAEVTATDLEVTGRIPDYLDGRYLRN
MAV_4505|M.avium_104 ---MTTTRTVA--NPYLDGFLAPVRAEVTAFDLPVTGRIPDHLDGRYLRN
Mb0673|M.bovis_AF2122/97 ---MTTAQAAESQNPYLEGFLAPVSTEVTATDLPVTGRIPEHLDGRYLRN
Rv0654|M.tuberculosis_H37Rv ---MTTAQAAESQNPYLEGFLAPVSTEVTATDLPVTGRIPEHLDGRYLRN
MAB_3874c|M.abscessus_ATCC_199 --MTATAQPAASTSPYLSGIWEPVQQEMDSVDLEVTGTLPAHLDGRYLRN
MSMEG_6224|M.smegmatis_MC2_155 MTDTDLTNLPGTGEFFRRGNYAPVPDELTDYDLPVTGAIPPELDGWYLRN
. : * *: * .* * * :* *** ****
Mflv_2111|M.gilvum_PYR-GCK GPNPIGDLDPALYHWFIGDGMVHGIRIRDGRAEWYRNRWVRGPAAARALG
TH_3959|M.thermoresistible__bu GPNPVGEIDPELYHWFVGDGMVHGIRLRDGRAEWYRNRWVRGPLTARALG
Mvan_4595|M.vanbaalenii_PYR-1 GPNPVAEVDPELYHWFTGDGMVHGIRIRDGKAEWYRNRWVRSPRVAEALG
MMAR_0993|M.marinum_M GPNPVAEVNPATYHWFTGDGMVHGVALRDGRASWYRNRWVRSMPVCAALG
MUL_0744|M.ulcerans_Agy99 GPNPVAEVNPATYHWFTGDGMVHGVALRDGRASWYRNRWVRSMPVCAALG
MAV_4505|M.avium_104 GPNPVAEVDPATYHWFSGDGMVHGVALRDGRARWYRNRWVRTAHVCAALG
Mb0673|M.bovis_AF2122/97 GPNPVAEVDPATYHWFTGDAMVHGVALRDGKARWYRNRWVRTPAVCAALG
Rv0654|M.tuberculosis_H37Rv GPNPVAEVDPATYHWFTGDAMVHGVALRDGKARWYRNRWVRTPAVCAALG
MAB_3874c|M.abscessus_ATCC_199 GPNPAAEVDPATYHLFSGDAMVHGLSLREGKAQWYRNRWVRTPSVSKTLG
MSMEG_6224|M.smegmatis_MC2_155 GPNPR----QATAHWFTGDGMIHGVRIENGAAKWYRNRWVRTDSFIESFP
**** * * **.*:**: :.:* * ******** ::
Mflv_2111|M.gilvum_PYR-GCK ESVPDG---RFPISGIGANTNVIGHAGKTLALIEAGVPNYELTDDLDTVG
TH_3959|M.thermoresistible__bu ERPPSE---ECSVSGIGANTNVIGHAGKTIALVESGVTNYELTEELDTVG
Mvan_4595|M.vanbaalenii_PYR-1 ETPPRG---DYGLTPIGANTNVIGHAGKTLALIEAGVAQYELTDDLDAVD
MMAR_0993|M.marinum_M ESPPSRLDPRAGMLGLGANTNVLSHAGRTLALVEGGVANYELTDDLDTVG
MUL_0744|M.ulcerans_Agy99 ESPPARRDPRAGMLGLGANTNVLSHAGRTLALVEGGVANYELTDDLDTVG
MAV_4505|M.avium_104 EPAPGGLDPRAGMLSVGPNTNVLSHAGQTLALVEGGGANYRLTEDLDTVG
Mb0673|M.bovis_AF2122/97 EPISARPHPRTGIIEGGPNTNVLTHAGRTLALVEAGVVNYELTDELDTVG
Rv0654|M.tuberculosis_H37Rv EPISARPHPRTGIIEGGPNTNVLTHAGRTLALVEAGVVNYELTDELDTVG
MAB_3874c|M.abscessus_ATCC_199 ETRPAAISANAGMGVIGPNTNVLGHAGRTLALVEGGIANYELTEDLDTIG
MSMEG_6224|M.smegmatis_MC2_155 VYNADG---TRNLRASVANTHIVNHAGKTLALVESSLP-YEITGDLETVG
. : .**::: ***:*:**:*.. *.:* :*:::.
Mflv_2111|M.gilvum_PYR-GCK TCDFDGTLSGGYTAHPKRDPDTGELHAVSYSMLRGNTVQYSVIDTGGRAR
TH_3959|M.thermoresistible__bu PCDFDGTLTGGYTAHPKRDPETGELHAVSYSMYRGNTVQYSVIGTDGRAR
Mvan_4595|M.vanbaalenii_PYR-1 VCDFDGTLAGGYTAHPKRDPGTGELHAVSYCLLNGNTVQYSVVGVDGRAR
MMAR_0993|M.marinum_M TCDFDGTLPGGYTAHPHRDPQTGELHAVSYSFTRGHTVQYSVIDTQGRAR
MUL_0744|M.ulcerans_Agy99 TCDFDGTLPGGYTAHPHRDPQTGELHAVSYSFTRGHMVQYSVIDTQGRAR
MAV_4505|M.avium_104 TCDFDGTLFGGYTAHPHRDPRTGELHAVSYSFGRGRRVQYSVIDTAGRAR
Mb0673|M.bovis_AF2122/97 PCDFDGTLHGGYTAHPQRDPHTGELHAVSYSFARGHRVQYSVIGTDGHAR
Rv0654|M.tuberculosis_H37Rv PCDFDGTLHGGYTAHPQRDPHTGELHAVSYSFARGHRVQYSVIGTDGHAR
MAB_3874c|M.abscessus_ATCC_199 TCDFDGTLPGGYTAHPHTDPDTGEMHAVSYSFARGSTVQYSVIDAHGRAR
MSMEG_6224|M.smegmatis_MC2_155 AYDFGGKLVDSMTAHPKICPTTGEMHFFGYGSLFEPYVTYHRADADGELT
**.*.* .. ****: * ***:* ..* * * .. *.
Mflv_2111|M.gilvum_PYR-GCK RTVDIEVAGSPMMHDFSLTERHVIFYDLPVTFDTRRAVE-MTVPRGLRLP
TH_3959|M.thermoresistible__bu RTVDIEVTGSPMMHDFSLTERHIVLYDLPVTFDARQAVS-MTVPRGLRLP
Mvan_4595|M.vanbaalenii_PYR-1 RTVDVEVTGSPMMHDFSLTERHVVFYDLPVTFDSRQAAT-IGVPRWLQLP
MMAR_0993|M.marinum_M RTVDIEVTGSPMMHDFSLTDKYVVIYDLPVTFDSAQVMP-VKMPSWLGVP
MUL_0744|M.ulcerans_Agy99 RTVDIEVTGSPMMHDFSLTDKYVVIYDLPVTFDSAQVMP-VKMPSWLGVP
MAV_4505|M.avium_104 RTVDIEVSGSPMMHDFSLTDEYVVIYDLPVTFDPVQVMP-ADVPRWLSAP
Mb0673|M.bovis_AF2122/97 RTVDIEVAGSPMMHSFSLTDNYVVIYDLPVTFDPMQVVP-ASVPRWLQRP
Rv0654|M.tuberculosis_H37Rv RTVDIEVAGSPMMHSFSLTDNYVVIYDLPVTFDPMQVVP-ASVPRWLQRP
MAB_3874c|M.abscessus_ATCC_199 RTVDIPVHGAPMMHDFTLTEKYVVFYDLPVTLDMSMISQSVPVPRLLRKP
MSMEG_6224|M.smegmatis_MC2_155 INRPVDVKAHTMMHDFAMTSGHVIFMDLPVVFDLDIALK-----------
. : * . .***.*::*. :::: ****.:*
Mflv_2111|M.gilvum_PYR-GCK ARLVLSALIGRVRIPDPVAARQPSGSGSDRRFPYSWNPRYPARIGVMPRD
TH_3959|M.thermoresistible__bu ARLVLSALIGRVRVPDPIAARQPRGNSSDRRFPYSWNPRYPARIGVMPRD
Mvan_4595|M.vanbaalenii_PYR-1 ARLVLSALIGRVRIPDPLAARRPGRDSSDAAFPYSWNPRYPARVGVMPRE
MMAR_0993|M.marinum_M ARLVMASLIGNVRVPSPIAAMMNRNPKPSNRLPYAWNPAYPARIGVMPRE
MUL_0744|M.ulcerans_Agy99 ARLVMASLIGNVRVPSLIAAMLNRNPKPSNRLPYAWNPAYPARIGVMPRE
MAV_4505|M.avium_104 ARLVVQSLLGRVQLPGPMATAMNRNRQPLHRMPYRWNDAYPARLGVMPRD
Mb0673|M.bovis_AF2122/97 ARLVIQSVLGRVRIPDPIAALGNRMQGHSDRLPYAWNPSYPARVGVMPRE
Rv0654|M.tuberculosis_H37Rv ARLVIQSVLGRVRIPDPIAALGNRMQGHSDRLPYAWNPSYPARVGVMPRE
MAB_3874c|M.abscessus_ATCC_199 AQIVMNSLIGKVKIPGPIAAKASQYSGKSPSIPYSWNPKYPARIGVMPRE
MSMEG_6224|M.smegmatis_MC2_155 ---------------------------GTGDMPYRWSDTYGARFGLLRRD
:** *. * **.*:: *:
Mflv_2111|M.gilvum_PYR-GCK GGSAD----VRWFDVEPCYVFHPMNAYDSP--ADDTIVLDVVRHPKMFDT
TH_3959|M.thermoresistible__bu GSNKD----VRWFDVEPCYVFHPMNAYDD----GDTIVLDVIRHPKMFDT
Mvan_4595|M.vanbaalenii_PYR-1 GGNGD----VRWFEVEPCYVFHPMNAFDD----GDTVVLDVVRHEKMFAT
MMAR_0993|M.marinum_M GGNRD----VRWFDIEPCYVYHPLNAYSEHRDGAEVLILDVVRYRRMFDR
MUL_0744|M.ulcerans_Agy99 GGNRD----VRWFDIEPCYVYHPLNAYSEHRDGAEVLILDVVRYRRMFDR
MAV_4505|M.avium_104 GGNDE----VRWFDIEPCYVYHPLNAYSEIRDGAKVLVLDVVRYSRMFDR
Mb0673|M.bovis_AF2122/97 GGNED----VRWFDIEPCYVYHPLNAYSECRNGAEVLVLDVVRYSRMFDR
Rv0654|M.tuberculosis_H37Rv GGNED----VRWFDIEPCYVYHPLNAYSECRNGAEVLVLDVVRYSRMFDR
MAB_3874c|M.abscessus_ATCC_199 GDDASRHAPVRWFEIDPCYVFHPLNGYSEMRNGAEVIVIDLVRYDSMFSQ
MSMEG_6224|M.smegmatis_MC2_155 DPFGE----IRWFDIDPCYIFHVVNAYEH----GNTVVVQAVRYPELWRD
. . :***:::***::* :*.:. ..:::: :*: ::
Mflv_2111|M.gilvum_PYR-GCK DHLGPN-EGPPTLERWTVDLVAGKVRESRVDDRAQEFPRVDERRVGKRHR
TH_3959|M.thermoresistible__bu DNLGPN-EGPPTLDRWTVDLADGKVRESRIDDRGQEFPRVDERLVGRRHR
Mvan_4595|M.vanbaalenii_PYR-1 DHLGPN-EGIPTLERWTVDLADGKVRESRIDDRGQEFPRVDERVVGRRHR
MMAR_0993|M.marinum_M DRRGPG-DTTPSLDRWVINLTTGAVSTECRDDRSQEFPRINETLLGKRHR
MUL_0744|M.ulcerans_Agy99 DRRGPG-DTSPSLDRWVINLTTGAVSTECRDDRSQEFPRINETLLGKRHR
MAV_4505|M.avium_104 DLRGPG-DTRPTLDRWTVNLATGAVGSELRDDRPQEFPRINEALLGRRHR
Mb0673|M.bovis_AF2122/97 DRRGPGGDSRPSLDRWTINLATGAVTAECRDDRAQEFPRINETLVGGPHR
Rv0654|M.tuberculosis_H37Rv DRRGPGGDSRPSLDRWTINLATGAVTAECRDDRAQEFPRINETLVGGPHR
MAB_3874c|M.abscessus_ATCC_199 DVLGPS-DAKGQLDRWTIDLAKGTVHMERKDDRHLEFPRINETFTGKKHR
MSMEG_6224|M.smegmatis_MC2_155 N---GSFDAQAVLWSWTIDLQTGAVTERQLDDLAVEFPRIDDRLAGLHAR
: . : * *.::* * * ** ****::: * *
Mflv_2111|M.gilvum_PYR-GCK YGYAP------CVGDGT-GDDELLKHDLVGGNSLTRAFGNGKVAGEFVFH
TH_3959|M.thermoresistible__bu YGYAP------TVGEGTSGGNVLLKHDFVGGNTQSRVFEPYHELGEFVFQ
Mvan_4595|M.vanbaalenii_PYR-1 YGYAP------TVTGGGSCADTLLKHDFVGTGSVARSFGAGKTLGEFVFH
MMAR_0993|M.marinum_M FGYTLGVDGGYLTGGTSDMSTSLYKQDYATGSSQVAPLDSDLLLGEMAFV
MUL_0744|M.ulcerans_Agy99 FGYTLGVDGGYLTGGTSDMSTSLYKQDYATGSSQVAPLDPDLLLGEMAFV
MAV_4505|M.avium_104 FGYTVGTDGGYLSDGASEMSTGLYKHDYATGSRRVAPLDPDLLIGEMCFV
Mb0673|M.bovis_AF2122/97 FAYTVGIEGGFLVGAGAALSTPLYKQDCVTGSSTVASLDPDLLIGEMVFV
Rv0654|M.tuberculosis_H37Rv FAYTVGIEGGFLVGAGAALSTPLYKQDCVTGSSTVASLDPDLLIGEMVFV
MAB_3874c|M.abscessus_ATCC_199 YGYLPDVEN--FFGSGEAAGASIVKYDYETGSTLSSRLDPAIVLGEMSFV
MSMEG_6224|M.smegmatis_MC2_155 YSVAAG-------------DHGWVRYDMTTGDAQRHDLGTG-GPGEAVFV
:. : * . : ** *
Mflv_2111|M.gilvum_PYR-GCK PSSP---DAAEDDGVLMGYVYDRTTDRSELAILDAATL--EDVATVKLPH
TH_3959|M.thermoresistible__bu PSSA---DAAEDDGVVMGYVYDRTTDRSELAILDAQTL--EDVARIRLPH
Mvan_4595|M.vanbaalenii_PYR-1 PSSP---DAAEDDGVLMGYIYDRPTDRSQLAILDAATL--DDVAAVNLPH
MMAR_0993|M.marinum_M PSPT---AAAEDDGVLMGYGYHRGSDEGQLVLLDAQTL--ELAACVHLPQ
MUL_0744|M.ulcerans_Agy99 PSPT---AAAEDHGVLMGYGYHRGSDEGQLVLLDAQTL--ELAACVHLPQ
MAV_4505|M.avium_104 PNPAGGQDGTEDDGILMGFGYHRGRDENQLVLLDAQSL--EQVAAVHLPQ
Mb0673|M.bovis_AF2122/97 PNPS---ARAEDDGILMGYGWHRGRDEGQLLLLDAQTL--ESIATVHLPQ
Rv0654|M.tuberculosis_H37Rv PNPS---ARAEDDGILMGYGWHRGRDEGQLLLLDAQTL--ESIATVHLPQ
MAB_3874c|M.abscessus_ATCC_199 PNPT---CAAEDDGVLIGFAVDRRSDEGQLLLLDATSL--DLMATVHLPQ
MSMEG_6224|M.smegmatis_MC2_155 PGAG---PADESNGWYLGYVYDPERDGSDLVILDASDFGGKPVARIELPR
*.. *..* :*: . * .:* :*** : . * :.**:
Mflv_2111|M.gilvum_PYR-GCK RVPSGFHGNWVPTA-
TH_3959|M.thermoresistible__bu RVPAGFHGNWVPSDG
Mvan_4595|M.vanbaalenii_PYR-1 RVPAGFHGNWVPTA-
MMAR_0993|M.marinum_M RVPLGFHGNWSAST-
MUL_0744|M.ulcerans_Agy99 RVPLGFHGNWSAST-
MAV_4505|M.avium_104 RVPMGFHGNWVPAG-
Mb0673|M.bovis_AF2122/97 RVPMGFHGNWAPTT-
Rv0654|M.tuberculosis_H37Rv RVPMGFHGNWAPTT-
MAB_3874c|M.abscessus_ATCC_199 RVPAGFHGNWAPR--
MSMEG_6224|M.smegmatis_MC2_155 RVPYGFHGNWISA--
*** ****** .