For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AELDATDERIVTLLMRNARATFAEIGEEVNLSAPAVKRRVDRLVGTGVIKGFTTVVDRAAVGWNTEAYVQ VYCQGTITPEALRRAWIDIPEVVSAATVTGTSDAMLRVLARDIQHLERALERIRASGDIDRTESIVVLSN LVDRAQVQG*
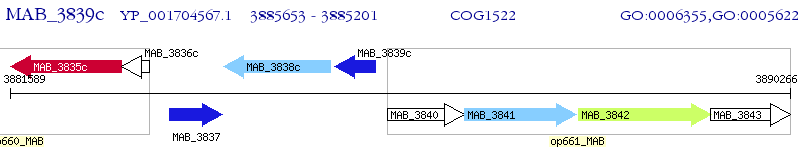
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3839c | - | - | 100% (150) | AsnC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3099c | - | 7e-14 | 32.87% (143) | AsnC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0892c | - | 9e-14 | 28.87% (142) | AsnC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2351 | - | 4e-56 | 71.72% (145) | AsnC family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_5069 | - | 4e-57 | 72.60% (146) | AsnC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2324 | - | 4e-56 | 71.72% (145) | AsnC family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3624 | - | 3e-56 | 71.14% (149) | AsnC family transcriptional regulator |
| M. avium 104 | MAV_2084 | - | 7e-57 | 73.10% (145) | AsnC-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1415 | - | 1e-57 | 71.43% (147) | AsnC-family protein transcriptional regulator |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1241 | - | 1e-56 | 71.14% (149) | AsnC family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1289 | - | 4e-59 | 76.03% (146) | AsnC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2351|M.bovis_AF2122/97 --------MDRLDDTDERILAELAEHARATFAEIGHKVSLSAPAVKRRVD
Rv2324|M.tuberculosis_H37Rv --------MDRLDDTDERILAELAEHARATFAEIGHKVSLSAPAVKRRVD
MAV_2084|M.avium_104 --------MDRLDDTDERILAELTEQARVTFAEIGDKVNLSAPAVKRRVD
MMAR_3624|M.marinum_M --------MDHLDDTDDRILVELTDHARATFAEIGQKVNLSAPAVKRRVD
MUL_1241|M.ulcerans_Agy99 --------MDHLDSTDDRILVELTDHARATFAEIGQKVNLSAPAVKRRVD
Mflv_5069|M.gilvum_PYR-GCK MFCCFRRMMDRLDDTDERILAELADNARATFAEIGQHVNLSAPAVKRRVD
Mvan_1289|M.vanbaalenii_PYR-1 --------MDRLDETDERILAELADNARATFAEIGQQVNLSAPAVKRRVD
MSMEG_1415|M.smegmatis_MC2_155 --------MPRVDETDERILGVLAEDARATYAEIGAQVNLSAPAVKRRVD
MAB_3839c|M.abscessus_ATCC_199 --------MAELDATDERIVTLLMRNARATFAEIGEEVNLSAPAVKRRVD
* .:* **:**: * .**.*:**** .*.***********
Mb2351|M.bovis_AF2122/97 RMLESGVIKGFTTVVDRNALGWNTEAYVQIFCHGRIAPDQLRAAWVNIPE
Rv2324|M.tuberculosis_H37Rv RMLESGVIKGFTTVVDRNALGWNTEAYVQIFCHGRIAPDQLRAAWVNIPE
MAV_2084|M.avium_104 RMLDSGVIKGFTTVVDRNALGWNTEAYVQVFCHGRIAPDELRTAWVDIPE
MMAR_3624|M.marinum_M RMIDNGVIKGFTTIVDRNALGWHTEAYVQVFCHGRIAPDQLQAAWIDIHE
MUL_1241|M.ulcerans_Agy99 RMIDNGVIKGFTTIVDRNALGWHTEAYVQVFCHGRIAPDQLQAAWIDIHE
Mflv_5069|M.gilvum_PYR-GCK RMLDNGVIRGFTTVIDRSALGWTTEAYVQVFCHGTIAPAELRAAWKDIPE
Mvan_1289|M.vanbaalenii_PYR-1 RMLDSGVIKGFTTVIDRSALGWTTEAFVQVFCHGTIAPTELRAAWKDIPE
MSMEG_1415|M.smegmatis_MC2_155 RMLADGVIRGFTTVVDRNVLGWNTEAYVQVFCHGTIAPDELRAAWVDIPE
MAB_3839c|M.abscessus_ATCC_199 RLVGTGVIKGFTTVVDRAAVGWNTEAYVQVYCQGTITPEALRRAWIDIPE
*:: ***:****::** .:** ***:**::*:* *:* *: ** :* *
Mb2351|M.bovis_AF2122/97 VVSAATVTGTSDAILHVLAHDMRHLEAALERIRSSADVERSESTVVLSNL
Rv2324|M.tuberculosis_H37Rv VVSAATVTGTSDAILHVLAHDMRHLEAALERIRSSADVERSESTVVLSNL
MAV_2084|M.avium_104 VFSAATVTGTPDAILHVLARDMRHLEVALERIRSSADVERSESIVVLSNL
MMAR_3624|M.marinum_M VVSAATVTGTSDAILHVLARDMGHLETALERIRSSADIERSESVVVLSNL
MUL_1241|M.ulcerans_Agy99 VVSAATVTGTSDAILHVLARDMGHLETALERIRSSADIERSESVVVLSNL
Mflv_5069|M.gilvum_PYR-GCK VVGAATVTGTADAMLHVLARDMRHLEEALERIRSSADIERSESIVVLSNI
Mvan_1289|M.vanbaalenii_PYR-1 VVSAATVTGTSDAMLHVLARDMRHLEEALERIRASADIERSESIVVLSNL
MSMEG_1415|M.smegmatis_MC2_155 VVGAATVTGTADAILHVLARDMRHLEEALERIRASADIERSESIVVLSNI
MAB_3839c|M.abscessus_ATCC_199 VVSAATVTGTSDAMLRVLARDIQHLERALERIRASGDIDRTESIVVLSNL
*..*******.**:*:***:*: *** ******:*.*::*:** *****:
Mb2351|M.bovis_AF2122/97 IDRMPP--
Rv2324|M.tuberculosis_H37Rv IDRMPP--
MAV_2084|M.avium_104 IDRMRP--
MMAR_3624|M.marinum_M IDRMRPQP
MUL_1241|M.ulcerans_Agy99 IDRMRPQP
Mflv_5069|M.gilvum_PYR-GCK IERSPG--
Mvan_1289|M.vanbaalenii_PYR-1 IERASR--
MSMEG_1415|M.smegmatis_MC2_155 IERARG--
MAB_3839c|M.abscessus_ATCC_199 VDRAQVQG
::*