For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STFRHIAGPVTYQFGSLAEVLAKASPPRSGDELAGCAAHSDAERAAARWVLAEVPLAAFLTEEIVPYDTD EVTRLILDSHDAAAFAVISHLTVGDFRDWLLETITKPHGAQILKEVSPGLTPEMVAAVSKLMRNQDLIAV GAAVRNHSAFRTTIGLPGTLATRLQPNHPTDDARGIAAATLDGLLLGCGDAVIGINPATDSPHAAGDLLR LIDDIRLRFDIPTQSCVLAHVTTTIELIERNLPVDLVFQSIAGTEGANESFGVNLALLREANEAGRSLSR GTVGNNVMYLETGQGSALSAGAHLGVGGVAVDQQTLEARAYAVAREVEPLLVNTVVGFIGPEYLYDGKQI IRAGLEDHFCGKLLGLPMGVDVCYTNHAEADSDDMDVLLTVLTAAGVAFVIAVPGADDVMLGYQSLSFHD ALFARRTFGLRPAPEFNDWLERMGMLERDGGLREIAVSDSPLRALI*
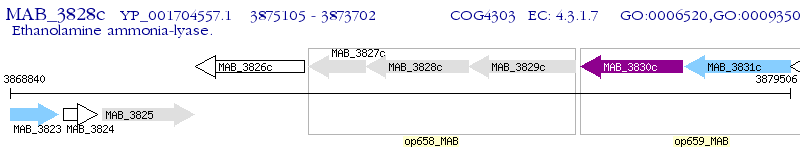
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3828c | - | - | 100% (467) | ethanolamine ammonia-lyase, large subunit |
| M. abscessus ATCC 19977 | MAB_2790c | - | 0.0 | 74.95% (463) | ethanolamine ammonia-lyase, heavy subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0664 | eutB | 0.0 | 73.49% (464) | ethanolamine ammonia-lyase large subunit, EutB |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1553 | eutB | 0.0 | 76.24% (463) | ethanolamine ammonia-lyase, large subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3197 | - | 0.0 | 76.04% (455) | ethanolamine ammonia lyase large subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0664|M.marinum_M MSYRQTISG-TTYAFDGLVDVLAKATPLRSGDQLAGCAAEHDAERAAAAW
Mvan_3197|M.vanbaalenii_PYR-1 MNYRQQVSG-HTYQFDGLVDLMAKASPPRSGDELAGCAAQSDAERAAAAW
MSMEG_1553|M.smegmatis_MC2_155 MRYRQQVSG-VTYTFDGLVEVMAKATPLRSGDQLAGCAAEHDGERAAAAW
MAB_3828c|M.abscessus_ATCC_199 MSTFRHIAGPVTYQFGSLAEVLAKASPPRSGDELAGCAAHSDAERAAARW
* : ::* ** *..*.:::***:* ****:******. *.***** *
MMAR_0664|M.marinum_M VLADMPLTTFLSDVVVPYETDEVTRLIIDSHDRQAFAPISHLTVGGLRDW
Mvan_3197|M.vanbaalenii_PYR-1 ILADLPLSLFLDEMVVPYETDDVTRLIVDTHDRDAFSSISHLTVGGFRDW
MSMEG_1553|M.smegmatis_MC2_155 VLADLPLDVFLNEELVPYDTDEVTRLIMDTHDRQAFSSVSHLTVGGFRDW
MAB_3828c|M.abscessus_ATCC_199 VLAEVPLAAFLTEEIVPYDTDEVTRLILDSHDAAAFAVISHLTVGDFRDW
:**::** ** : :***:**:*****:*:** **: :******.:***
MMAR_0664|M.marinum_M LLDTAARDDSAARIAAIAPGLTPEMVAAVCKIMRNQDLILVAAATTATAA
Mvan_3197|M.vanbaalenii_PYR-1 LLDTASGPRSAERIAAVASGLTPEMVAAVSKIMRNQDLISVAAATTVSAG
MSMEG_1553|M.smegmatis_MC2_155 LLDVAAEDDSAARLAAVAPGLTPEMVAAVSKIMRNQDLIAVSAAARVTAA
MAB_3828c|M.abscessus_ATCC_199 LLETITKPHGAQILKEVSPGLTPEMVAAVSKLMRNQDLIAVGAAVRNHSA
**:. : .* : ::.**********.*:******* *.**. :.
MMAR_0664|M.marinum_M FRTTIGLPGRLATRLQPNHPTDDPRGIAAGMLDGLLMGCGDAVVGINPAT
Mvan_3197|M.vanbaalenii_PYR-1 FRTSIGVPGTLATRLQPNHPTDDPRGIAAATLDGLLMGCGDAVIGINPAT
MSMEG_1553|M.smegmatis_MC2_155 LRTTVGIPGTMATRLQPNHPTDDPRGIAAAVLDGLLLGCGDAVIGINPAT
MAB_3828c|M.abscessus_ATCC_199 FRTTIGLPGTLATRLQPNHPTDDARGIAAATLDGLLLGCGDAVIGINPAT
:**::*:** :************.*****. *****:******:******
MMAR_0664|M.marinum_M DSPQATADLLHLLDDIRQRFEIPMQSCVLCHVTTTTELIDKGVPVDLVFQ
Mvan_3197|M.vanbaalenii_PYR-1 DSPRATSDLLHLLDEIRQRFDIPMQSCVLTHVTTTMELIERGAPVDLVFQ
MSMEG_1553|M.smegmatis_MC2_155 DSPQATADLLYLLDGIRTRYDIPAQSCVLSHITTTIGLIEAGAPVDLVFQ
MAB_3828c|M.abscessus_ATCC_199 DSPHAAGDLLRLIDDIRLRFDIPTQSCVLAHVTTTIELIERNLPVDLVFQ
***:*:.*** *:* ** *::** ***** *:*** **: . *******
MMAR_0664|M.marinum_M SIAGTEGANSSFGVNIPMLLEANEAARSLGRGTVGDNVMYLETGQGSALS
Mvan_3197|M.vanbaalenii_PYR-1 SIAGTQGANASFGVTLPMLREANEAGRALGRGTVGDNVMYLETGQGSALS
MSMEG_1553|M.smegmatis_MC2_155 SIAGTEGANTAFGVNLQLLREGNEAARSLNRGTVGNNVMYLETGQGSALS
MAB_3828c|M.abscessus_ATCC_199 SIAGTEGANESFGVNLALLREANEAGRSLSRGTVGNNVMYLETGQGSALS
*****:*** :***.: :* *.***.*:*.*****:**************
MMAR_0664|M.marinum_M AGAHLGTGGKPVDQQTLEARAYAVARTLQPLLINTVVGFIGPEYLYDGKQ
Mvan_3197|M.vanbaalenii_PYR-1 AGAHLGTGGRPVDQQTLETRAYAVARALDPLLVNTVVGFIGPEYLYDGKQ
MSMEG_1553|M.smegmatis_MC2_155 SGAHLGVGGKPVDQQTLEARAYAVARDLSPLLVNTVVGFIGPEYLYDGKQ
MAB_3828c|M.abscessus_ATCC_199 AGAHLGVGGVAVDQQTLEARAYAVAREVEPLLVNTVVGFIGPEYLYDGKQ
:*****.** .*******:******* :.***:*****************
MMAR_0664|M.marinum_M IIRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQDDMDTLLTLLGVAGAAF
Mvan_3197|M.vanbaalenii_PYR-1 IVRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQDDMDTLLTLLGVAGAAF
MSMEG_1553|M.smegmatis_MC2_155 IIRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQNDMDTLLMLLAAAGVAF
MAB_3828c|M.abscessus_ATCC_199 IIRAGLEDHFCGKLLGLPMGVDVCYTNHAEADSDDMDVLLTVLTAAGVAF
*:******************************.:***.** :* .**.**
MMAR_0664|M.marinum_M VIAVPGADDIMLGYQSLSFHDPLYVRQVLRLRPAPEFEAWLTGLGMADAN
Mvan_3197|M.vanbaalenii_PYR-1 VITVPGADDVMLGYQSLSFHDALYVRKALNLRPAPEFEAWLARLGMADGD
MSMEG_1553|M.smegmatis_MC2_155 VITVPGADDVMLGYQSLSFHDVLVARRTLGLRPAPEFETWLRKVGMVDDA
MAB_3828c|M.abscessus_ATCC_199 VIAVPGADDVMLGYQSLSFHDALFARRTFGLRPAPEFNDWLERMGMLERD
**:******:*********** * .*:.: *******: ** :** :
MMAR_0664|M.marinum_M GRILPIDLATSPLRALATGA--
Mvan_3197|M.vanbaalenii_PYR-1 GRVLPVDPAGSPLLALAAGE--
MSMEG_1553|M.smegmatis_MC2_155 GRLTPFDVSRSPLRELTVAEAS
MAB_3828c|M.abscessus_ATCC_199 GGLREIAVSDSPLRALI-----
* : . : *** *