For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KYRQQVAGVNYAFDGLVDVMAKATPLRSGDELAGCAAGSDAERAAAAWVLADLPLDTFLNEAVVPYESDE VTRLIIDSHDRGAYSAVSHLTVGGFRDWLLESASRDDGARRIAEVSAGITPEMAAAVSKIMRNQDLIAVG AAMHVSAAFRTTIGGKGTLATRLQPNHPTDDPRGVAAAVLDGLLLGCGDAVIGINPATDSPEATGDLLKL LDSIRSRYDIPTQSCVLSHITTTIGLIERGAPVDLTFQSIAGTEGANSAFGVDIALLREGRDATRALRRG TVGDNVMYLETGQGSALSSRAHLGAGGKPVDQQTLEARAYAVARDLEPFLVNTVVGFIGPEYLYDGKQIV RAGLEDHFCGKLLGLPMGVDVCYTNHAEADQNDMDTLLILLAAAGVAFVITVPGADDVMLGYQSLSFHDV LQARRTLGLRPAPEFEQWLHAIGMVDDDGRLTPFDVTSSPLRALTISGAR*
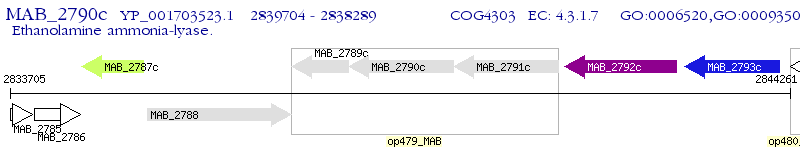
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2790c | - | - | 100% (471) | ethanolamine ammonia-lyase, heavy subunit |
| M. abscessus ATCC 19977 | MAB_3828c | - | 0.0 | 74.95% (463) | ethanolamine ammonia-lyase, large subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0664 | eutB | 0.0 | 76.34% (465) | ethanolamine ammonia-lyase large subunit, EutB |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1553 | eutB | 0.0 | 83.62% (470) | ethanolamine ammonia-lyase, large subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3197 | - | 0.0 | 78.28% (465) | ethanolamine ammonia lyase large subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0664|M.marinum_M MSYRQTISGTTYAFDGLVDVLAKATPLRSGDQLAGCAAEHDAERAAAAWV
Mvan_3197|M.vanbaalenii_PYR-1 MNYRQQVSGHTYQFDGLVDLMAKASPPRSGDELAGCAAQSDAERAAAAWI
MAB_2790c|M.abscessus_ATCC_199 MKYRQQVAGVNYAFDGLVDVMAKATPLRSGDELAGCAAGSDAERAAAAWV
MSMEG_1553|M.smegmatis_MC2_155 MRYRQQVSGVTYTFDGLVEVMAKATPLRSGDQLAGCAAEHDGERAAAAWV
* *** ::* .* *****:::***:* ****:****** *.*******:
MMAR_0664|M.marinum_M LADMPLTTFLSDVVVPYETDEVTRLIIDSHDRQAFAPISHLTVGGLRDWL
Mvan_3197|M.vanbaalenii_PYR-1 LADLPLSLFLDEMVVPYETDDVTRLIVDTHDRDAFSSISHLTVGGFRDWL
MAB_2790c|M.abscessus_ATCC_199 LADLPLDTFLNEAVVPYESDEVTRLIIDSHDRGAYSAVSHLTVGGFRDWL
MSMEG_1553|M.smegmatis_MC2_155 LADLPLDVFLNEELVPYDTDEVTRLIMDTHDRQAFSSVSHLTVGGFRDWL
***:** **.: :***::*:*****:*:*** *::.:*******:****
MMAR_0664|M.marinum_M LDTAARDDSAARIAAIAPGLTPEMVAAVCKIMRNQDLILVAAATTATAAF
Mvan_3197|M.vanbaalenii_PYR-1 LDTASGPRSAERIAAVASGLTPEMVAAVSKIMRNQDLISVAAATTVSAGF
MAB_2790c|M.abscessus_ATCC_199 LESASRDDGARRIAEVSAGITPEMAAAVSKIMRNQDLIAVGAAMHVSAAF
MSMEG_1553|M.smegmatis_MC2_155 LDVAAEDDSAARLAAVAPGLTPEMVAAVSKIMRNQDLIAVSAAARVTAAL
*: *: .* *:* ::.*:****.***.********* *.** .:*.:
MMAR_0664|M.marinum_M RTTIGLPGRLATRLQPNHPTDDPRGIAAGMLDGLLMGCGDAVVGINPATD
Mvan_3197|M.vanbaalenii_PYR-1 RTSIGVPGTLATRLQPNHPTDDPRGIAAATLDGLLMGCGDAVIGINPATD
MAB_2790c|M.abscessus_ATCC_199 RTTIGGKGTLATRLQPNHPTDDPRGVAAAVLDGLLLGCGDAVIGINPATD
MSMEG_1553|M.smegmatis_MC2_155 RTTVGIPGTMATRLQPNHPTDDPRGIAAAVLDGLLLGCGDAVIGINPATD
**::* * :***************:**. *****:******:*******
MMAR_0664|M.marinum_M SPQATADLLHLLDDIRQRFEIPMQSCVLCHVTTTTELIDKGVPVDLVFQS
Mvan_3197|M.vanbaalenii_PYR-1 SPRATSDLLHLLDEIRQRFDIPMQSCVLTHVTTTMELIERGAPVDLVFQS
MAB_2790c|M.abscessus_ATCC_199 SPEATGDLLKLLDSIRSRYDIPTQSCVLSHITTTIGLIERGAPVDLTFQS
MSMEG_1553|M.smegmatis_MC2_155 SPQATADLLYLLDGIRTRYDIPAQSCVLSHITTTIGLIEAGAPVDLVFQS
**.**.*** *** ** *::** ***** *:*** **: *.****.***
MMAR_0664|M.marinum_M IAGTEGANSSFGVNIPMLLEANEAARSLGRGTVGDNVMYLETGQGSALSA
Mvan_3197|M.vanbaalenii_PYR-1 IAGTQGANASFGVTLPMLREANEAGRALGRGTVGDNVMYLETGQGSALSA
MAB_2790c|M.abscessus_ATCC_199 IAGTEGANSAFGVDIALLREGRDATRALRRGTVGDNVMYLETGQGSALSS
MSMEG_1553|M.smegmatis_MC2_155 IAGTEGANTAFGVNLQLLREGNEAARSLNRGTVGNNVMYLETGQGSALSS
****:***::*** : :* *..:* *:* *****:**************:
MMAR_0664|M.marinum_M GAHLGTGGKPVDQQTLEARAYAVARTLQPLLINTVVGFIGPEYLYDGKQI
Mvan_3197|M.vanbaalenii_PYR-1 GAHLGTGGRPVDQQTLETRAYAVARALDPLLVNTVVGFIGPEYLYDGKQI
MAB_2790c|M.abscessus_ATCC_199 RAHLGAGGKPVDQQTLEARAYAVARDLEPFLVNTVVGFIGPEYLYDGKQI
MSMEG_1553|M.smegmatis_MC2_155 GAHLGVGGKPVDQQTLEARAYAVARDLSPLLVNTVVGFIGPEYLYDGKQI
****.**:********:******* *.*:*:******************
MMAR_0664|M.marinum_M IRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQDDMDTLLTLLGVAGAAFV
Mvan_3197|M.vanbaalenii_PYR-1 VRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQDDMDTLLTLLGVAGAAFV
MAB_2790c|M.abscessus_ATCC_199 VRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQNDMDTLLILLAAAGVAFV
MSMEG_1553|M.smegmatis_MC2_155 IRAGLEDHFCGKLLGLPMGVDVCYTNHAEADQNDMDTLLMLLAAAGVAFV
:*******************************:****** **..**.***
MMAR_0664|M.marinum_M IAVPGADDIMLGYQSLSFHDPLYVRQVLRLRPAPEFEAWLTGLGMADANG
Mvan_3197|M.vanbaalenii_PYR-1 ITVPGADDVMLGYQSLSFHDALYVRKALNLRPAPEFEAWLARLGMADGDG
MAB_2790c|M.abscessus_ATCC_199 ITVPGADDVMLGYQSLSFHDVLQARRTLGLRPAPEFEQWLHAIGMVDDDG
MSMEG_1553|M.smegmatis_MC2_155 ITVPGADDVMLGYQSLSFHDVLVARRTLGLRPAPEFETWLRKVGMVDDAG
*:******:*********** * .*:.* ******** ** :**.* *
MMAR_0664|M.marinum_M RILPIDLATSPLRALATGA--
Mvan_3197|M.vanbaalenii_PYR-1 RVLPVDPAGSPLLALAAGE--
MAB_2790c|M.abscessus_ATCC_199 RLTPFDVTSSPLRALTISGAR
MSMEG_1553|M.smegmatis_MC2_155 RLTPFDVSRSPLRELTVAEAS
*: *.* : *** *: .