For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTENAQPRLKTRYREEIKTALNDEFKYANVMQIPGVVKVVVNMGVGDAARDAKLINGAVTDLALITGQK PEIRKARKSIAQFKLREGMPIGARVTLRGDRMWEFLDRLVSIALPRIRDFRGLSPKQFDGKGNYTFGLTE QSMFHEIDVDSIDRPRGMDITVVTSATTDDEGRALLRQLGFPFKEN*
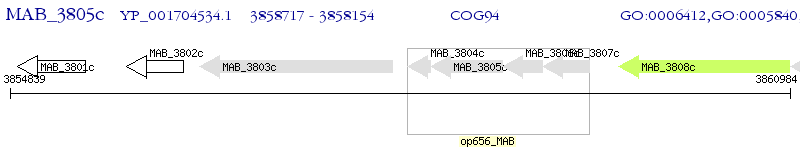
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3805c | rplE | - | 100% (187) | 50S ribosomal protein L5 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0737 | rplE | 3e-87 | 82.89% (187) | 50S ribosomal protein L5 |
| M. gilvum PYR-GCK | Mflv_5035 | rplE | 3e-90 | 88.83% (188) | 50S ribosomal protein L5 |
| M. tuberculosis H37Rv | Rv0716 | rplE | 3e-87 | 82.89% (187) | 50S ribosomal protein L5 |
| M. leprae Br4923 | MLBr_01847 | rplE | 1e-89 | 86.10% (187) | 50S ribosomal protein L5 |
| M. marinum M | MMAR_1047 | rplE | 2e-85 | 82.11% (190) | 50S ribosomal protein L5, RplE |
| M. avium 104 | MAV_4453 | rplE | 1e-89 | 86.10% (187) | 50S ribosomal protein L5 |
| M. smegmatis MC2 155 | MSMEG_1467 | rplE | 8e-94 | 89.84% (187) | 50S ribosomal protein L5 |
| M. thermoresistible (build 8) | TH_0311 | rplE | 6e-93 | 89.84% (187) | PROBABLE 50S RIBOSOMAL PROTEIN L5 RPLE |
| M. ulcerans Agy99 | MUL_0806 | rplE | 6e-85 | 81.58% (190) | 50S ribosomal protein L5 |
| M. vanbaalenii PYR-1 | Mvan_1334 | rplE | 2e-90 | 88.83% (188) | 50S ribosomal protein L5 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5035|M.gilvum_PYR-GCK MTS--TETKTLPRLKQRYREEIREQLLKEFGYANVMQIPGVVKVVVNMGV
Mvan_1334|M.vanbaalenii_PYR-1 MTT--TETKTLPRLKQRYREEIRDQLLKEFGYANVMQIPGVTKVVVNMGV
TH_0311|M.thermoresistible__bu MTT--AE-KTLPRLKQRYREEIREALQKEFGYTNVMQIPTVVKVVVNMGV
MSMEG_1467|M.smegmatis_MC2_155 MTT--TE-KALPRLKQRYREEIREALQQEFNYANVMQIPGVVKVVVNMGV
MAB_3805c|M.abscessus_ATCC_199 MTT--TE-NAQPRLKTRYREEIKTALNDEFKYANVMQIPGVVKVVVNMGV
Mb0737|M.bovis_AF2122/97 MTT---AQKVQPRLKERYRSEIRDALRKQFGYGNVMQIPTVTKVVVNMGV
Rv0716|M.tuberculosis_H37Rv MTT---AQKVQPRLKERYRSEIRDALRKQFGYGNVMQIPTVTKVVVNMGV
MMAR_1047|M.marinum_M MSTTDSAVKIPPRLKERYRSEIRDVLHKQFGYGNVMQIPTVTKVVVNMGV
MUL_0806|M.ulcerans_Agy99 MSTTDSAVKIPPRLKERYRSEIRDVLHKQFGYGNVMQIPTVTKVVVNMGV
MLBr_01847|M.leprae_Br4923 MST---AEKVQPRLKQRYRSEIREALNKQFSYGNVMQIPTVVKVVVNMGV
MAV_4453|M.avium_104 MTT---TEKVQPRLKERYRNEIRDSLQQQFGYANVMQIPTVTKVVVNMGI
*:: : **** ***.**: * .:* * ****** *.*******:
Mflv_5035|M.gilvum_PYR-GCK GDAARDAKLINGAVNDLMLITGQKPEIRKARKSIAQFKLREGMPIGARVT
Mvan_1334|M.vanbaalenii_PYR-1 GDAARDAKLINGAVNDLALITGQKPEIRRARKSIAQFKLREGMPIGARVT
TH_0311|M.thermoresistible__bu GDAARDAKLINGAVNDLALITGQKPEIRRARKSIAQFKLREGMPIGARAT
MSMEG_1467|M.smegmatis_MC2_155 GDAARDAKLINGAINDLALITGQKPEVRRARKSIAQFKLREGMPIGARVT
MAB_3805c|M.abscessus_ATCC_199 GDAARDAKLINGAVTDLALITGQKPEIRKARKSIAQFKLREGMPIGARVT
Mb0737|M.bovis_AF2122/97 GEAARDAKLINGAVNDLALITGQKPEVRRARKSIAQFKLREGMPVGVRVT
Rv0716|M.tuberculosis_H37Rv GEAARDAKLINGAVNDLALITGQKPEVRRARKSIAQFKLREGMPVGVRVT
MMAR_1047|M.marinum_M GEAARDAKLINGAVSDLALITGQRPEIRKARKSIAQFKLREGMPVGVRAT
MUL_0806|M.ulcerans_Agy99 GEAARDAKLINGAVSDLALITGQRPEIRKARKSIAQFKLREGMPVGVRAT
MLBr_01847|M.leprae_Br4923 GDAARDAKLINGAVSDLALITGQKPEVRKARKSIAQFKLREGMPIGVRVT
MAV_4453|M.avium_104 GEAARDAKLINGAVNDLALITGQRPEIRRARKSIAQFKLREGMPIGARVT
*:***********:.** *****:**:*:***************:*.*.*
Mflv_5035|M.gilvum_PYR-GCK LRGDRMWEFLDRLISISLPRIRDFRGLSGKQFDGTGNYTFGLTEQSVFHE
Mvan_1334|M.vanbaalenii_PYR-1 LRGDRMWEFLDRLISISLPRIRDFRGLSGKQFDGTGNYTFGLTEQSVFHE
TH_0311|M.thermoresistible__bu LRGDRMWEFLDRLISIALPRIRDFRGLSPKQFDGTGNYTFGLTEQSVFHE
MSMEG_1467|M.smegmatis_MC2_155 LRGDRMWEFLDRLISIALPRIRDFRGLSPKQFDGTGNYTFGLNEQSMFHE
MAB_3805c|M.abscessus_ATCC_199 LRGDRMWEFLDRLVSIALPRIRDFRGLSPKQFDGKGNYTFGLTEQSMFHE
Mb0737|M.bovis_AF2122/97 LRGDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQAVFHE
Rv0716|M.tuberculosis_H37Rv LRGDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQAVFHE
MMAR_1047|M.marinum_M LRGDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQSVFHE
MUL_0806|M.ulcerans_Agy99 LRGDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQPVFHE
MLBr_01847|M.leprae_Br4923 LRDDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQSVFHE
MAV_4453|M.avium_104 LRGDRMWEFLDRLTSIALPRIRDFRGLSPKQFDGVGNYTFGLAEQSVFHE
**.********** **:*********** ***** ******* **.:***
Mflv_5035|M.gilvum_PYR-GCK IDVDSIDRPRGMDITVVTSATNDDEGRALLRALGFPFKEN
Mvan_1334|M.vanbaalenii_PYR-1 IDVDSIDRPRGMDITVVTSATNDDEGRALLRALGFPFKEN
TH_0311|M.thermoresistible__bu IDVDSIDRPRGMDITVVTSATTDDEGRALLRALGFPFKEN
MSMEG_1467|M.smegmatis_MC2_155 IDVDSIDRPRGMDITVVTTATNDAEGRALLRALGFPFKEN
MAB_3805c|M.abscessus_ATCC_199 IDVDSIDRPRGMDITVVTSATTDDEGRALLRQLGFPFKEN
Mb0737|M.bovis_AF2122/97 VDVDKIDRVRGMDINVVTSAATDDEGRALLRALGFPFKEN
Rv0716|M.tuberculosis_H37Rv VDVDKIDRVRGMDINVVTSAATDDEGRALLRALGFPFKEN
MMAR_1047|M.marinum_M IDVDKIDRVRGMDINVVTSATTDEEGRALLRALGFPFKEN
MUL_0806|M.ulcerans_Agy99 IDVDKIDRVRGMDINVVTSATTDEEGRALLRALGFPFKEN
MLBr_01847|M.leprae_Br4923 IDVDKIDRVRGMDINVVTSATTDDEGRALLRALGFPFKEN
MAV_4453|M.avium_104 IDVDKIDRVRGMDINVVTSATTDDEGRALLRALGFPFKEN
:***.*** *****.***:*:.* ******* ********