For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GPVSTTSTTTDGLPPSTGATTAQAVIDLGAIAHNVKLLREHAGTARLMTVVKADGYGHGAVRVARAALAA GADELGVTSIAEALQIRAAGLDAPLLAWLHAPGADYAAALHADVEVAVSSLGQLGELLAAARSTGRQAIT SIKVDTGLNRNGVPAGYTRDLFEALAKAQAEESIRLRGIMSHFAYADQPGHPTVDMQIGRFNEALELARG FGLRYELAHLSNSAATLTRPDARYDMVRPGIAVYGINPLGAAESATSNRGRTSFDLIPAMSLTCAVSSVK PVSAGEGVSYGHVWTAPRDTNAALIGIGYADGVVRALGNRFEVAIGGRRYPNIGRVCMDQFVVDLGDNDA GVQAGDVATLFGPGTGGEPTAQDWAELLDTIAYEVVTGPRGRVVRTYRDSRAVE*
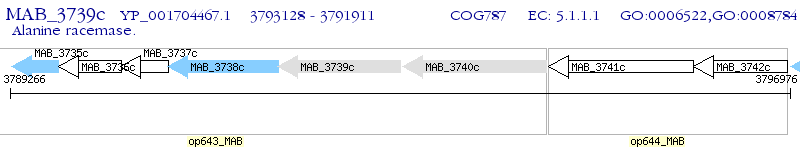
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3739c | alr | - | 100% (405) | alanine racemase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3457c | alr | 1e-129 | 58.23% (407) | alanine racemase |
| M. gilvum PYR-GCK | Mflv_4934 | alr | 1e-120 | 58.62% (377) | alanine racemase |
| M. tuberculosis H37Rv | Rv3423c | alr | 1e-129 | 58.23% (407) | alanine racemase |
| M. leprae Br4923 | MLBr_00375 | alr | 1e-127 | 59.01% (383) | alanine racemase |
| M. marinum M | MMAR_1119 | alr | 1e-129 | 59.28% (388) | alanine racemase, Alr |
| M. avium 104 | MAV_4372 | alr | 1e-128 | 58.51% (388) | alanine racemase |
| M. smegmatis MC2 155 | MSMEG_1575 | alr | 1e-117 | 56.30% (389) | alanine racemase |
| M. thermoresistible (build 8) | TH_1694 | - | 1e-129 | 59.70% (397) | ALANINE RACEMASE ALR |
| M. ulcerans Agy99 | MUL_0879 | alr | 1e-129 | 59.54% (388) | alanine racemase |
| M. vanbaalenii PYR-1 | Mvan_1484 | alr | 1e-127 | 60.00% (395) | alanine racemase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00375|M.leprae_Br4923 ----------------------MAVTPISLRPGVLAEAVVDLGAIDYNVR
MAV_4372|M.avium_104 ----------------------MAVTPISLTPGVLAEALVDLGAIEHNVR
Mb3457c|M.bovis_AF2122/97 MKRFWENVGKPNDTTDGRGTTSLAMTPISQTPGLLAEAMVDLGAIEHNVR
Rv3423c|M.tuberculosis_H37Rv MKRFWENVGKPNDTTDGRGTTSLAMTPISQTPGLLAEAMVDLGAIEHNVR
MMAR_1119|M.marinum_M ----------------------MTTTPISLTPGVLAEAVVDLGAIAHNVR
MUL_0879|M.ulcerans_Agy99 ----------------------MTTTPISLTPGVLAEAVVDLGAIAHNVR
Mflv_4934|M.gilvum_PYR-GCK --------------------MTTT----ENRLMNTPSAVIDLDAIAHNVG
Mvan_1484|M.vanbaalenii_PYR-1 --------------------MTTTPTMTQKLPQALPTASIDLDAIAHNVA
MSMEG_1575|M.smegmatis_MC2_155 --------------------MQTTEPMTPPAPLASAQTVIDLGAIDHNVR
TH_1694|M.thermoresistible__bu -----------------MGSDTTGHPAQRTVSSATAEAVVDLGAIAHNVR
MAB_3739c|M.abscessus_ATCC_199 -------------MGPVSTTSTTTDGLPPSTGATTAQAVIDLGAIAHNVK
. : :**.** :**
MLBr_00375|M.leprae_Br4923 VLREHAGMAQLMVVLKADAYGHGATQVALAALAAGAAELGVATVDEALAL
MAV_4372|M.avium_104 LLCEQARGAQVMAVVKADGYGHGAVQTARAALAAGAAELGVATVDEALAL
Mb3457c|M.bovis_AF2122/97 VLREHAGHAQLMAVVKADGYGHGATRVAQTALGAGAAELGVATVDEALAL
Rv3423c|M.tuberculosis_H37Rv VLREHAGHAQLMAVVKADGYGHGATRVAQTALGAGAAELGVATVDEALAL
MMAR_1119|M.marinum_M LLRERAGSAQVMAVVKADGYGHGATAVARTALAAGAVELGVASVDEALTL
MUL_0879|M.ulcerans_Agy99 LLRERAGSAQVMAVVKADGYGHGATAVARTALAAGAVELGVASVDEALTL
Mflv_4934|M.gilvum_PYR-GCK VLRDRAGRAAVMAVVKADGYGHGATAVARAALAAGATNLGVATLDEALSL
Mvan_1484|M.vanbaalenii_PYR-1 VLREHAGPAAVMAVVKADGYGHGATQVARAALAAGAAELGVATVGEALAL
MSMEG_1575|M.smegmatis_MC2_155 VLRELAGSADVMAVVKADAYGHGALPVARTALAAGAAALGVATIPEALAL
TH_1694|M.thermoresistible__bu VVRERAGSAQVMAVVKADGYGHGATAVARTALAAGATELGVATVGEALAL
MAB_3739c|M.abscessus_ATCC_199 LLREHAGTARLMTVVKADGYGHGAVRVARAALAAGADELGVTSIAEALQI
:: : * * :*.*:***.***** .* :**.*** ***::: *** :
MLBr_00375|M.leprae_Br4923 RADGISAPVLAWLHPPGIDFGPALLADVQIAVSSVRQLDELLDAVRRTGR
MAV_4372|M.avium_104 RAAGISAPVLAWLHPPGIDFRPALLAGVQIGLSSQRQLDELLTAVRDTGR
Mb3457c|M.bovis_AF2122/97 RADGITAPVLAWLHPPGIDFGPALLADVQVAVSSLRQLDELLHAVRRTGR
Rv3423c|M.tuberculosis_H37Rv RADGITAPVLAWLHPPGIDFGPALLADVQVAVSSLRQLDELLHAVRRTGR
MMAR_1119|M.marinum_M RADGITAPVLAWLHAPGMDFGPALAADVQIAISSIRQLDEVLDAARRTGT
MUL_0879|M.ulcerans_Agy99 RADGITAPVLAWLHAPGMDFGPALAADVQIAISSIRQLDEALDAARRTGT
Mflv_4934|M.gilvum_PYR-GCK RRDGVTAPVLSWLHPPGTDFTPAVQADIEVALSSRRQLDDLLRAVARTGR
Mvan_1484|M.vanbaalenii_PYR-1 RRDGVTARVLAWLHPPGTDFAPALLADVDVAVSSLRQFTDLLDAAGRTGR
MSMEG_1575|M.smegmatis_MC2_155 REGGITAPVLAWLHPPGTDFAPAIAADVEVAVSSRRQLEQVTAAAAEVGR
TH_1694|M.thermoresistible__bu RRDGVTAPVLSWLNPPGTDYAPALRAQIEVAVSSRRQLDEVLDAARRTGV
MAB_3739c|M.abscessus_ATCC_199 RAAGLDAPLLAWLHAPGADYAAALHADVEVAVSSLGQLGELLAAARSTGR
* *: * :*:**:.** *: .*: * :::.:** *: : *. .*
MLBr_00375|M.leprae_Br4923 TATVTVKADTGLNRNGVVTDQYPAMLTALQRAVVEDAVRLRGLMSHLVYA
MAV_4372|M.avium_104 TATVTVKVDTGLNRNGVPPAQYPSMLTALRRAVAEQAIVPRGLMSHMVYA
Mb3457c|M.bovis_AF2122/97 TATVTVKVDTGLNRNGVGPAQFPAMLTALRQAMAEDAVRLRGLMSHMVYA
Rv3423c|M.tuberculosis_H37Rv TATVTVKVDTGLNRNGVGPAQFPAMLTALRQAMAEDAVRLRGLMSHMVYA
MMAR_1119|M.marinum_M TATVTVKIDTGLNRNGVAPALYPEMVTRLRQAVAEDAIRLRGLMTHMVHA
MUL_0879|M.ulcerans_Agy99 TATVTVKIDTGLNRNGVAPALYPEMVTRLRQAVAEDAIRLRGLMTHMVHA
Mflv_4934|M.gilvum_PYR-GCK TATVTVKADTGLSRNGVSPADHPGLVSALARAQTDGAVTVHGLMSHLAHG
Mvan_1484|M.vanbaalenii_PYR-1 TADVTVKADTGLSRNGVGEADYPALVDALRRAASDGAVRVRGLMSHLAHG
MSMEG_1575|M.smegmatis_MC2_155 TATVTVKVDTGLSRNGVGAADYPEVLDVLRRAQADGAIRVRGLMSHLVHG
TH_1694|M.thermoresistible__bu TATLSVKVDTGLNRNGVSRAEYPEVLAAVRRAQAEGAIRLRAVMSHLVAA
MAB_3739c|M.abscessus_ATCC_199 QAITSIKVDTGLNRNGVPAGYTRDLFEALAKAQAEESIRLRGIMSHFAYA
* ::* ****.**** :. : :* : :: :.:*:*:. .
MLBr_00375|M.leprae_Br4923 DQPDNPSNDVQGKRFAALLAQAHEQGLRFEVAHLSNSSATMSRPDLAYDL
MAV_4372|M.avium_104 DQPANPVNDVQAQRFTDMLAQAREQGVRFEVAHLSNSSATMSRPDLAFDM
Mb3457c|M.bovis_AF2122/97 DKPDDSINDVQAQRFTAFLAQAREQGVRFEVAHLSNSSATMARPDLTFDL
Rv3423c|M.tuberculosis_H37Rv DKPDDSINDVQAQRFTAFLAQAREQGVRFEVAHLSNSSATMARPDLTFDL
MMAR_1119|M.marinum_M DAPEKPINDIQSQRFKQMFDHARDQGVRFEVAHLSNSSATMARPDLTLDL
MUL_0879|M.ulcerans_Agy99 DAPEKPINDIQSQRFKQMLDHARDQGVRFEVAHLSNSSATMARPDLTLDL
Mflv_4934|M.gilvum_PYR-GCK DVPDHPFNDEQARRLTEMASEARSAGVAYDVVHLCNSPAALTRPDLAFDM
Mvan_1484|M.vanbaalenii_PYR-1 DDPEHPFNDVQARRLTEMAKLAGARGVSYDVVHLSNSPAALTRPDLAFDM
MSMEG_1575|M.smegmatis_MC2_155 DDPENPFNGLQGQRLADMRVYAREHGVDYEVAHLCNSPAAMTRPDLAFEM
TH_1694|M.thermoresistible__bu DEPDNPVTDIQRQRFSEMRAQARDAGLRFEIAHLCNSAGTMSRPDLAFDM
MAB_3739c|M.abscessus_ATCC_199 DQPGHPTVDMQIGRFNEALELARGFGLRYELAHLSNSAATLTRPDARYDM
* * .. . * *: * *: :::.**.**..:::*** ::
MLBr_00375|M.leprae_Br4923 VRPGIAVYGLSPV-------PSRG--DMGLIPAMTVKCAVAMVKSIRAGE
MAV_4372|M.avium_104 VRPGIAVYGLSPV-------PELG--DMGLVPAMTVKCTVALVKSIRAGE
Mb3457c|M.bovis_AF2122/97 VRPGIAVYGLSPV-------PALG--DMGLVPAMTVKCAVALVKSIRAGE
Rv3423c|M.tuberculosis_H37Rv VRPGIAVYGLSPV-------PALG--DMGLVPAMTVKCAVALVKSIRAGE
MMAR_1119|M.marinum_M VRPGIAVYGLSPV-------PRLG--DMGLVPAMTVKCAVALVKSVSAGE
MUL_0879|M.ulcerans_Agy99 VRPGIAVYGLSPV-------PRLG--DMGLVPAMTVKCAVALVKSVSAGE
Mflv_4934|M.gilvum_PYR-GCK VRPGIAVYGQTPI-------PERG--DMGLRPAMTVKCPVALVRSVRAGD
Mvan_1484|M.vanbaalenii_PYR-1 VRPGIAVYGQTPI-------PQRG--DFGLRPAMTVKCPVSAVRPVRAGD
MSMEG_1575|M.smegmatis_MC2_155 VRPGISLYGLSPI-------PERG--DMGLRPAMTLKCPVALVRSVHAGD
TH_1694|M.thermoresistible__bu VRAGIAVYGLSPI-------PQRG--DLGLKPAMTLKCPVALVKQVRAGE
MAB_3739c|M.abscessus_ATCC_199 VRPGIAVYGINPLGAAESATSNRGRTSFDLIPAMSLTCAVSSVKPVSAGE
**.**::** .*: . * .:.* ***::.*.*: *: : **:
MLBr_00375|M.leprae_Br4923 GVSYGHDWIAQHDTNLALLPVGYADGVFRSLGGRLDVLINGKRRPGVGRI
MAV_4372|M.avium_104 SVSYGHTWTAQRDTNLALLPVGYADGIFRSLGGRLQVSINGRRRPGVGRI
Mb3457c|M.bovis_AF2122/97 GVSYGHTWIAPRDTNLALLPIGYADGVFRSLGGRLEVLINGRRCPGVGRI
Rv3423c|M.tuberculosis_H37Rv GVSYGHTWIAPRDTNLALLPIGYADGVFRSLGGRLEVLINGRRCPGVGRI
MMAR_1119|M.marinum_M GVSYGHTWIAPHDTNVALLPIGYADGVFRSLGGRLEVLINGKRRPGVGRV
MUL_0879|M.ulcerans_Agy99 GVSYGHTWIAPHDTNVALLPIGYADGVFRSLGGRLEVLINGKRRPGVGRV
Mflv_4934|M.gilvum_PYR-GCK GVSYGHTWLAPRDTTLALIPAGYADGVFRALSNRFEVTINGRRYPNVGRV
Mvan_1484|M.vanbaalenii_PYR-1 AVSYGHTWTAQTDTTLALVPAGYADGVFRALSNRFEVSINGRRYPNVGRI
MSMEG_1575|M.smegmatis_MC2_155 GVSYGHRWVADRDTTLGLLPIGYADGVYRALSGRIDVLIKGRRRRAVGRI
TH_1694|M.thermoresistible__bu GVSYGHAWTADRDTTIALLPIGYADGVFRPLSNRFEVLINGRRHRNVGRV
MAB_3739c|M.abscessus_ATCC_199 GVSYGHVWTAPRDTNAALIGIGYADGVVRALGNRFEVAIGGRRYPNIGRV
.***** * * **. .*: *****: *.*..*::* * *:* :**:
MLBr_00375|M.leprae_Br4923 CMDQFVVDLGPGPIDVAEGDEAILFGPGARGEPTAQDWADLLGTIHYEVV
MAV_4372|M.avium_104 CMDQFVVDLGPGRPDVAEGDEAILFGPGSNGEPTAQDWADLLGTIHYEVV
Mb3457c|M.bovis_AF2122/97 CMDQFMVDLGPGPLDVAEGDEAILFGPGIRGEPTAQDWADLVGTIHYEVV
Rv3423c|M.tuberculosis_H37Rv CMDQFMVDLGPGPLDVAEGDEAILFGPGIRGEPTAQDWADLVGTIHYEVV
MMAR_1119|M.marinum_M CMDQFLVDLGPGPLDVAEGDEAILFGPGTRGEPTAQDWADLVGTIHYEVV
MUL_0879|M.ulcerans_Agy99 CMDQFLVDLGPGPLDVAEGDEAILFGPGTRGEPTAQDWADLVGTIHYEVV
Mflv_4934|M.gilvum_PYR-GCK CMDQFVVDLGPGPVDVAEGDDAILFGPGDDGEPTAQDWADLLGTINYEVV
Mvan_1484|M.vanbaalenii_PYR-1 CMDQFVVDLGPGPVDVAEGDDAILFGPGTDGEPTAQDWAELLDTINYEVV
MSMEG_1575|M.smegmatis_MC2_155 CMDQFVVDLGPDADDVAVGDDAILFGPGANGEPTAQDWAELLDTIHYEVV
TH_1694|M.thermoresistible__bu CMDQFVVDLGPGPVDVTEGDEAILFGPGDSGEPSAQDWADLLDTIHYEVV
MAB_3739c|M.abscessus_ATCC_199 CMDQFVVDLGDNDAGVQAGDVATLFGPGTGGEPTAQDWAELLDTIAYEVV
*****:**** . .* ** * ***** ***:*****:*:.** ****
MLBr_00375|M.leprae_Br4923 TSLRGRITRTYREAQTVDR-------------
MAV_4372|M.avium_104 TSPRGRITRTYREAHTVES-------------
Mb3457c|M.bovis_AF2122/97 TSPRGRITRTYREAENR---------------
Rv3423c|M.tuberculosis_H37Rv TSPRGRITRTYREAENR---------------
MMAR_1119|M.marinum_M TSPRGRITRVYREAETVER-------------
MUL_0879|M.ulcerans_Agy99 TSPRGRITRVYREAETVER-------------
Mflv_4934|M.gilvum_PYR-GCK TSPRGRIVRGYVGGAQ----------------
Mvan_1484|M.vanbaalenii_PYR-1 TSPRGRIVRTYVGGKGGPQ-------------
MSMEG_1575|M.smegmatis_MC2_155 TSPRGRVTRTYLPAGQQD--------------
TH_1694|M.thermoresistible__bu TSPRGRVVRTYREADARAAAPRHAGRSEVGSV
MAB_3739c|M.abscessus_ATCC_199 TGPRGRVVRTYRDSRAVE--------------
*. ***:.* * .