For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ANTYAFDMPSIIATKRDGAELSPAAIDWLIGQYTSGDIGEEQVAALLMAIYLRGMNGAETAAWTAAILAS GERLDLSGLSRPTVDKHSTGGVGDKISLPLVAVVAACGAAVPQLSGRGLGHTGGTLDKLESVAGIRVDLS NDQIRQQLSDVGAVICAAGASLAPADRKLYALRDITGTVESIPLIAGSIMGKKLAEGTAALVLDVKVGAG AFMKSEQRARELASAMVDLGTAHGVATSALLTAMDTPLGATAGNAVEVQESLDVLAGGGPEDVVALTVTL AREMLAAAGIDGKDPAATLTDGTAMDVFRTMIHAQGGDLAVPLPLGQCQETVLAPVGGVMRRIDAMPVGI AAWRLGAGRSRPGEAVQHGAGVRIHRRPGEAVVAGEPLFTLYTDTQERLPAALDALEGGWEIGAAPTATP LIIDRLPV*
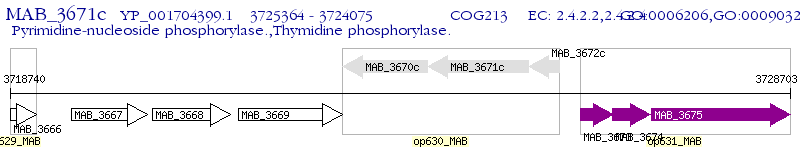
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3671c | deoA | - | 100% (429) | thymidine phosphorylase |
| M. abscessus ATCC 19977 | MAB_1970 | trpD | 2e-07 | 26.75% (157) | anthranilate phosphoribosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3343c | deoA | 1e-160 | 68.63% (424) | thymidine phosphorylase |
| M. gilvum PYR-GCK | Mflv_4842 | deoA | 1e-155 | 66.28% (430) | thymidine phosphorylase |
| M. tuberculosis H37Rv | Rv3314c | deoA | 1e-160 | 68.63% (424) | thymidine phosphorylase |
| M. leprae Br4923 | MLBr_00883 | trpD | 5e-12 | 28.21% (156) | anthranilate phosphoribosyltransferase |
| M. marinum M | MMAR_1205 | deoA | 1e-157 | 68.07% (429) | thymidine phosphorylase DeoA |
| M. avium 104 | MAV_4295 | deoA | 1e-164 | 70.14% (422) | thymidine phosphorylase |
| M. smegmatis MC2 155 | MSMEG_1675 | deoA | 1e-151 | 67.48% (412) | thymidine phosphorylase |
| M. thermoresistible (build 8) | TH_2179 | deoA | 1e-155 | 65.42% (428) | PROBABLE THYMIDINE PHOSPHORYLASE DEOA (TDRPASE) (PYRIMIDINE |
| M. ulcerans Agy99 | MUL_1365 | deoA | 1e-158 | 68.07% (429) | thymidine phosphorylase |
| M. vanbaalenii PYR-1 | Mvan_1591 | deoA | 1e-159 | 67.60% (429) | thymidine phosphorylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4842|M.gilvum_PYR-GCK ------MSFDAAGPISHLDAPSVIRTKRDGGALSDEAIDWVIDAYTRGEV
Mvan_1591|M.vanbaalenii_PYR-1 ------------MTQFTFDMPSIIRTKRDGGVLSDDAIDWVIDAYTHGRV
MSMEG_1675|M.smegmatis_MC2_155 -------------------------------MLSDDAIDWVIDNYTRGRV
TH_2179|M.thermoresistible__bu ---VTRATFHASGAGSAFDAPTVIRIKRDGGVLSDEAIDWVIDAYTHGRI
Mb3343c|M.bovis_AF2122/97 ------------MTDFAFDAPTVIRTKRDGGRLSDAAIDWVVKAYTDGRV
Rv3314c|M.tuberculosis_H37Rv ------------MTDFAFDAPTVIRTKRDGGRLSDAAIDWVVKAYTDGRV
MMAR_1205|M.marinum_M ------------MTDFEFDAPTVIRIKRDGGRLSDAAIDWVIDAYTEGRV
MUL_1365|M.ulcerans_Agy99 ------------MTDFEFDAPTVIRIKRDGGRLSDAAIDWVIDAYTEGRV
MAV_4295|M.avium_104 -------------MRPGFDAPTVIRTKRDGGRLSDAAIDWVIDAYTRGLV
MAB_3671c|M.abscessus_ATCC_199 -----------MANTYAFDMPSIIATKRDGAELSPAAIDWLIGQYTSGDI
MLBr_00883|M.leprae_Br4923 MVLSSQPPSSRSATGSVLSWPQVLGRLTAGQDLTRGQAAWAMGQVMTGNA
*: * : *
Mflv_4842|M.gilvum_PYR-GCK AEAQMSALLMAIFLRGMTGHEIARWTAAMIASGERLDFSDLRRDGKPLAL
Mvan_1591|M.vanbaalenii_PYR-1 AEAQMSALLMAIFLRGMTNGEIARWTAAMVDSGERLDFSDLRRDGKPLAL
MSMEG_1675|M.smegmatis_MC2_155 ADEQMSALLMAIFLRGMTPGEITRWTAAMVNSGERFDFTDLERDGRPLAL
TH_2179|M.thermoresistible__bu ADEQMSALLMAIFLRGMTGPEIARWTSAMVASGERFDFTDLRRDGRPLAL
Mb3343c|M.bovis_AF2122/97 ADEQMSALLMAIVWRGMDRGEIARWTAAMLASGARLDFTDL-----PLAT
Rv3314c|M.tuberculosis_H37Rv ADEQMSALLMAIVWRGMDRGEIARWTAAMLASGARLDFTDL-----PLAT
MMAR_1205|M.marinum_M APEQMSALLMAIVLQGMDRGETARWTSAMLASGPRLDFSDLRAGGAPLVT
MUL_1365|M.ulcerans_Agy99 APEQMSALLMAIVLQGMDRGETARWTSAMLASGPRLDFSDLRAGGAPLVT
MAV_4295|M.avium_104 AEEQMAALLMAILLRGMDGGETAAWTSAMLASGDRLDFSDLG-----VPT
MAB_3671c|M.abscessus_ATCC_199 GEEQVAALLMAIYLRGMNGAETAAWTAAILASGERLDLSGLSR-----PT
MLBr_00883|M.leprae_Br4923 RPAQIAAFAVAMKMKVPTAAEVSELADVMLSHARKLPTEAICQDIG--ET
*::*: :*: : * : : .:: . :: :
Mflv_4842|M.gilvum_PYR-GCK VDKHSTGGVGD---KITIPLVPVVMACGGAVPQAAGRGLGHTGGTLDKLE
Mvan_1591|M.vanbaalenii_PYR-1 VDKHSTGGVGD---KITIPLVSVVMACGGAVPQAAGRGLGHTGGTLDKLE
MSMEG_1675|M.smegmatis_MC2_155 VDKHSTGGVGD---KLTIPLVPVVIACGAAVPQAAGRGLGHTGGTLDKLE
TH_2179|M.thermoresistible__bu VDKHSTGGVGD---KITIPLVPVVMACGGAVPQAAGRGLGHTGGTLDKLE
Mb3343c|M.bovis_AF2122/97 VDKHSTGGVGD---KITLPLVPVVAACGGAVPQASGRGLGHTGGTLDKLE
Rv3314c|M.tuberculosis_H37Rv VDKHSTGGVGD---KITLPLVPVVAACGGAVPQASGRGLGHTGGTLDKLE
MMAR_1205|M.marinum_M VDKHSTGGVGD---KITLPLVPVVVACGGTVPQASGRGLGHTGGTLDKLE
MUL_1365|M.ulcerans_Agy99 VDKHSTGGVGD---KITLPLVPVVVACGGTVPQASGRGLGHTGGTLDKLE
MAV_4295|M.avium_104 VDKHSTGGVGD---KITLPLVPVVAACGAAVPQASGRGLGHTGGTLDKLD
MAB_3671c|M.abscessus_ATCC_199 VDKHSTGGVGD---KISLPLVAVVAACGAAVPQLSGRGLGHTGGTLDKLE
MLBr_00883|M.leprae_Br4923 VDIAGTGGDGANTVNLSTMAAIVVAAAGVPVVKHGNRASSSLSGGADTLE
** .*** * ::: . ** *.* .* : ..*. . .* *.*:
Mflv_4842|M.gilvum_PYR-GCK SIPGFTAEITKSQIRQQLCDLGAAIFAAG-ELAPADRKIYALRDVTATTE
Mvan_1591|M.vanbaalenii_PYR-1 SIPGFTAEISKTQIRQQLCELGAAIFAAG-ELAPADRKIYALRDVTATTE
MSMEG_1675|M.smegmatis_MC2_155 AIPGFSAELTKTQIRQQLCGVGAAIFAAG-ELAPADRKIYALRDVTATTE
TH_2179|M.thermoresistible__bu SIPGFSAELTQAQIRRQLEEIGGAIFAAG-ELAPADRKMYALRDVTATVD
Mb3343c|M.bovis_AF2122/97 SITGFTANLSNQRVREQLCDVGAAIFAAG-QLAPADAKLYALRDITGTVE
Rv3314c|M.tuberculosis_H37Rv SITGFTANLSNQRVREQLCDVGAAIFAAG-QLAPADAKLYALRDITGTVE
MMAR_1205|M.marinum_M SISGFTATLSNQQVREQLCDVGAAIFAAG-ELAPADAKLYALRDITGTVE
MUL_1365|M.ulcerans_Agy99 SISGLTATLSNQQVREQLCDVGAAIFAAG-ELAPADAKLYALRDITGTVE
MAV_4295|M.avium_104 AIAGFSAQLPSRRVREQLREVGAAIFAAG-DLAPADAKLYALRDITATVE
MAB_3671c|M.abscessus_ATCC_199 SVAGIRVDLSNDQIRQQLSDVGAVICAAGASLAPADRKLYALRDITGTVE
MLBr_00883|M.leprae_Br4923 ALG-VRIDLGPEQVANSLAEIGIGFCFAP-LFQPSYRYASAVRHEIG---
:: . : :: ..* :* : * : *: *:*. .
Mflv_4842|M.gilvum_PYR-GCK SLPLIASSVMSKKIAEGTKALVLDSKVGSGAFLDSEAESRELARTMVELG
Mvan_1591|M.vanbaalenii_PYR-1 SLPLIASSVMSKKIAEGTRALVLDTKVGSGAFLKTEAESRELARTMVELG
MSMEG_1675|M.smegmatis_MC2_155 SLPLIASSIMSKKLAEGARALVLDVKVGRGAFLKTEAESRELARTMVELG
TH_2179|M.thermoresistible__bu SLPLIASSVMSKKIAEGTRALVLDTKVGSGAFLKTEAEARELARTMIDLG
Mb3343c|M.bovis_AF2122/97 SLPLIASSIMSKKLAEGAGALVLDVKVGSGAFMRSPVQARELAHTMVELG
Rv3314c|M.tuberculosis_H37Rv SLPLIASSIMSKKLAEGAGALVLDVKVGSGAFMRSPVQARELAHTMVELG
MMAR_1205|M.marinum_M SLPLIASSIMSKKLAEGAGALVLDVKVGSGAFLKSETQSRELAHTMVELG
MUL_1365|M.ulcerans_Agy99 SLPLIASSIMSKKLAEGAGALVLDVKVGSGAFLKSETQSRELAHTMVELG
MAV_4295|M.avium_104 SLPLIASSVMSKKLAEGAGALVLDVKVGSGALVKTEEQCRDLAHTMVGLG
MAB_3671c|M.abscessus_ATCC_199 SIPLIAGSIMGKKLAEGTAALVLDVKVGAGAFMKSEQRARELASAMVDLG
MLBr_00883|M.leprae_Br4923 -VPTVFNILGPLTNPAWPRAGLIGCAFGN------------LAEVMAGVF
:* : . : . . . * ::. .* ** .* :
Mflv_4842|M.gilvum_PYR-GCK VEHGVQTRALLTDMQTPLGRTVGNAVEVVESLEVLAGGGPDDVVELTLAL
Mvan_1591|M.vanbaalenii_PYR-1 TAHGVRTRALLTDMNTPLGRTVGNAVEVAESLEVLAGGGPDDVVELTLAL
MSMEG_1675|M.smegmatis_MC2_155 NDYGVPTRALLTDMNRPLGLSVGNAVEVAESLDVLAGGGPADVVELTVAL
TH_2179|M.thermoresistible__bu TAHGLVTRALLTDMHRPLGAAVGNAVEVAESVEVLAGGGPSDVVELTLAL
Mb3343c|M.bovis_AF2122/97 AAHGVPTRALLTEMNCPLGRTVGNALEVAEALEVLAGGGPPDVVELTLRL
Rv3314c|M.tuberculosis_H37Rv AAHGVPTRALLTEMNCPLGRTVGNALEVAEALEVLAGGGPPDVVELTLRL
MMAR_1205|M.marinum_M AAHGVPTRALLTDMNCPLGSTVGNALEVAEALEVLDGGGPPDVVELTVRL
MUL_1365|M.ulcerans_Agy99 AAHGVPTRALLTDMNCPLGSTVGNALEVAEALEVLDGGGPPDVVELTVRL
MAV_4295|M.avium_104 AAHGVPTRALLTDMNRPLGATVGNALEVAEALEVLAGGGPPDVVELTLRL
MAB_3671c|M.abscessus_ATCC_199 TAHGVATSALLTAMDTPLGATAGNAVEVQESLDVLAGGGPEDVVALTVTL
MLBr_00883|M.leprae_Br4923 AAR----RSSVLVVHGDDGLDELTTTTTSTIWRVQAG----TVDKLTFDP
: : :. * .: . * * * **.
Mflv_4842|M.gilvum_PYR-GCK AREMCDVAGLDGVDPAQTLRDGTAMDRFRDLVAAQGGDVDSLDAAMLPLG
Mvan_1591|M.vanbaalenii_PYR-1 AREMCDAAGLDGVDPAETLRDGTAMDRFRALVAAQGGDPD----AALPLG
MSMEG_1675|M.smegmatis_MC2_155 AREMLAAAGIDGVDPAETLRDGSAMDRFRALVAAQGGDLS----LPLPIG
TH_2179|M.thermoresistible__bu AAEMLDAAGIDGEDPADTLRDGSAMDAYRALINAQGGDPD----ASLPRG
Mb3343c|M.bovis_AF2122/97 AGEMLELAGIHGRDPAQTLRDGTAMDRFRWLVAAQGGDLS----KPLPIG
Rv3314c|M.tuberculosis_H37Rv AGEMLELAGIHGRDPAQTLRDGTAMDRFRRLVAAQGGDLS----KPLPIG
MMAR_1205|M.marinum_M AAEMLDLVGIDGLDPAETLRDGSAMDRFRRLIAAQGGELA----KPLPIG
MUL_1365|M.ulcerans_Agy99 AAEMLDLVGIDGRDPAETLRDGSAMDRFRRLIAAQGGELA----KPLPIG
MAV_4295|M.avium_104 AAEMLALAGVGDRDPADTLHDGTAMDRFRRLVAAQGGDLS----VPLPIG
MAB_3671c|M.abscessus_ATCC_199 AREMLAAAGIDGKDPAATLTDGTAMDVFRTMIHAQGGDLA----VPLPLG
MLBr_00883|M.leprae_Br4923 VEFGFARAHLDDLLGGDAQTN---AAKVRAVLAGAKG----------PVR
. . : . . : : * :: . * *
Mflv_4842|M.gilvum_PYR-GCK QHTETISAPRSGTMGDIDAMAVGLAVWRLGAGRSVPGEQVQFGAGMRIHR
Mvan_1591|M.vanbaalenii_PYR-1 AHSETVSAPRGGTMGDIDAMAVGLAVWRLGAGRSAPGEQVQFGAGMRIHR
MSMEG_1675|M.smegmatis_MC2_155 AASETVTAPRGGVMGDIDAMGVAMAAWRLGAGRAQPGEPVQLGAGVRLHR
TH_2179|M.thermoresistible__bu AHTETVTAPRSGTMGDLDAMAVGMAVWRLGAGRSAPGEQVQHGAGLRIHR
Mb3343c|M.bovis_AF2122/97 SHSETVTAGASGTMGDIDAMAVGLAAWRLGAGRSRPGARVQHGAGVRIHR
Rv3314c|M.tuberculosis_H37Rv SHSETVTAGASGTMGDIDAMAVGLAAWRLGAGRSRPGARVQHGAGVRIHR
MMAR_1205|M.marinum_M AHSETVRAGRGGTMGDIDAMAVGLAAWRLGAGRSRPGARVQHGAGIRIHR
MUL_1365|M.ulcerans_Agy99 AHSETVRAGRGGTMGDIDAMAVGLAAWRLGAGRSRPGARVQHGAGIRIHR
MAV_4295|M.avium_104 RHSETVTAPRGGTMGDIDAMAVGLAAWRLGAGRSRPGERVQAGAGLRIHR
MAB_3671c|M.abscessus_ATCC_199 QCQETVLAPVGGVMRRIDAMPVGIAAWRLGAGRSRPGEAVQHGAGVRIHR
MLBr_00883|M.leprae_Br4923 DAVVLNAAGAIVAYAGLSSHAEWSPAWDDGLARAS--AAIDSGAAEQLLA
* . :.: ..* * .*: :: **. ::
Mflv_4842|M.gilvum_PYR-GCK KPGEAVAAGEPLFTLYTETPHRLAAAASELDGAWSVGDHAPPTRPLIIDR
Mvan_1591|M.vanbaalenii_PYR-1 RPGEPVAAGEPLFTLYTDTPERLAGAVSELDGAWSVGDEPPARRPLIIDR
MSMEG_1675|M.smegmatis_MC2_155 RPGEPVAAGETLFTLYTDTPDRLPAALGELDGAWSVGDAPPQELPLIIDR
TH_2179|M.thermoresistible__bu RPGDPVAAGEPLFTLYTDTPERFEAAIAELDGGFSVGDTAPDRRPLIIDR
Mb3343c|M.bovis_AF2122/97 RPGEPVVVGEPLFTLYTNAPERFGAARAELAGGWSIRDSPPQVRPLIVDR
Rv3314c|M.tuberculosis_H37Rv RPGEPVVVGEPLFTLYTNAPERFGAARAELAGGWSIRDSPPQVRPLIVDR
MMAR_1205|M.marinum_M RPGEPVAAGEPLFTLYTDTPERFGAAVAELAAGFSVADSAPTTRPLIIDR
MUL_1365|M.ulcerans_Agy99 RPGEPVAAGEPLFTLYTDTPERFGAAVAELAAGFSVADSAPTTRPLIIDR
MAV_4295|M.avium_104 RPGEPVAAGEPLFTLHTDTPERFGAALAELDGGWNVGDGAPAPRPLIIDR
MAB_3671c|M.abscessus_ATCC_199 RPGEAVVAGEPLFTLYTDTQERLPAALDALEGGWEIG-AAPTATPLIIDR
MLBr_00883|M.leprae_Br4923 R-----------WVRFSQQV------------------------------
: :. .::
Mflv_4842|M.gilvum_PYR-GCK ISM
Mvan_1591|M.vanbaalenii_PYR-1 ITG
MSMEG_1675|M.smegmatis_MC2_155 ID-
TH_2179|M.thermoresistible__bu VV-
Mb3343c|M.bovis_AF2122/97 IV-
Rv3314c|M.tuberculosis_H37Rv IV-
MMAR_1205|M.marinum_M ISR
MUL_1365|M.ulcerans_Agy99 ISR
MAV_4295|M.avium_104 IVT
MAB_3671c|M.abscessus_ATCC_199 LPV
MLBr_00883|M.leprae_Br4923 ---