For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATLTFVYADSTAVVGPLATSSEPHSWDLCAQHASRITAPRGWELVRYNGPLPSNPEDDDLVALADAVRET TGGVRVAAAGFSEPALDVSAPAPNVAPRPVHPAPVGRRRGHLRVLPDPSE*
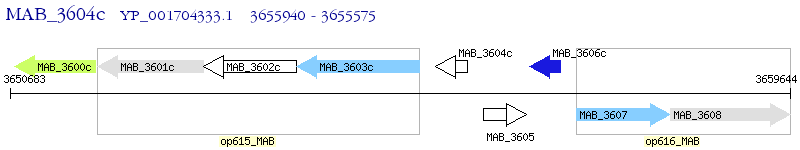
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3604c | - | - | 100% (121) | hypothetical protein MAB_3604c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3286c | - | 6e-39 | 61.83% (131) | hypothetical protein Mb3286c |
| M. gilvum PYR-GCK | Mflv_4727 | - | 3e-40 | 64.62% (130) | hypothetical protein Mflv_4727 |
| M. tuberculosis H37Rv | Rv3258c | - | 6e-39 | 61.83% (131) | hypothetical protein Rv3258c |
| M. leprae Br4923 | MLBr_00762 | - | 1e-41 | 65.41% (133) | hypothetical protein MLBr_00762 |
| M. marinum M | MMAR_1284 | - | 2e-39 | 60.45% (134) | hypothetical protein MMAR_1284 |
| M. avium 104 | MAV_4221 | - | 9e-39 | 59.70% (134) | hypothetical protein MAV_4221 |
| M. smegmatis MC2 155 | MSMEG_1833 | - | 1e-42 | 68.55% (124) | hypothetical protein MSMEG_1833 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2603 | - | 2e-39 | 60.45% (134) | hypothetical protein MUL_2603 |
| M. vanbaalenii PYR-1 | Mvan_1736 | - | 2e-41 | 68.29% (123) | hypothetical protein Mvan_1736 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3286c|M.bovis_AF2122/97 -------------------MRVSGASAALVHDSLSVVNVPRRCCRPGCPH
Rv3258c|M.tuberculosis_H37Rv -------------------MRVSGASAALVHDSLSVVNVPRRCCRPGCPH
MMAR_1284|M.marinum_M -------------------MRVSAPSAAFPHDSLCLVNVPRRCCRPGCPH
MUL_2603|M.ulcerans_Agy99 -------------------MRVSAPSAAFPHDSLCLVNVPRRCCRPGCPH
MAV_4221|M.avium_104 --------------------------------------------------
MLBr_00762|M.leprae_Br4923 -------------------MRVSGASATFSHDSLSVVNVPRRCCRPGCPH
Mflv_4727|M.gilvum_PYR-GCK MLAPGHGHDRNGWASAAPWLRVSGVMQAIVHANLLLVNVPRRCCRPGCPH
Mvan_1736|M.vanbaalenii_PYR-1 ------------------------------------MNVPRRCCRPGCPH
MSMEG_1833|M.smegmatis_MC2_155 ------------------------------------MNVPRRCCRPGCPH
MAB_3604c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3286c|M.bovis_AF2122/97 YAVATLTFVYSDSTAVIGPLATAREPHSWDLCVGHAGRITAPRGWELVRH
Rv3258c|M.tuberculosis_H37Rv YAVATLTFVYSDSTAVIGPLATAREPHSWDLCVGHAGRITAPRGWELVRH
MMAR_1284|M.marinum_M YAVATLTFVYSDSTAVIGPLATAREPHSWDLCVGHAGRITAPRGWELVRH
MUL_2603|M.ulcerans_Agy99 YAVATLTFVYSDSTAVIGPLATAREPHSWDLCVGHAGRITAPRGWELVRH
MAV_4221|M.avium_104 --MATLTFVYSDSTAVVGPLATAREPHSWDLCVNHAGRITAPRGWDLVRH
MLBr_00762|M.leprae_Br4923 YAVATLTFVYSDSTAVVGPLATVREPHSWDLCVDHAARITAPRGWELVRH
Mflv_4727|M.gilvum_PYR-GCK YAVATLTFVYADSTAVVGPLATVSEPHSWDLCVMHAGRITAPRGWELVRH
Mvan_1736|M.vanbaalenii_PYR-1 YAVATLTFVYSDSTAVVGPLATVSEPHSWDLCVMHAGRITAPRGWELVRH
MSMEG_1833|M.smegmatis_MC2_155 YAVATLTFVYSDSTAVVGPLATVSEPHSWDLCVGHASRITAPKGWELVRH
MAB_3604c|M.abscessus_ATCC_199 --MATLTFVYADSTAVVGPLATSSEPHSWDLCAQHASRITAPRGWELVRY
:*******:*****:***** ********. **.*****:**:***:
Mb3286c|M.bovis_AF2122/97 AGPLPSHPDEDDLVALADAVREGGPSAGRRHHPGGNGAPLHGFDDFP---
Rv3258c|M.tuberculosis_H37Rv AGPLPSHPDEDDLVALADAVREGGPSAGRRHHPGGNGAPLHGFDDFP---
MMAR_1284|M.marinum_M AGPLPTHPDEDDLVALADAVREQGPTAD-ASCAGATGAAHNGFSDQVMRH
MUL_2603|M.ulcerans_Agy99 AGPLPTHPDEDDLVALADAVREQGPTAD-ASCAGATGAAHNGFSDQVMRH
MAV_4221|M.avium_104 AGPLPTHPDEDDLVALADAVREGGSAERTLPYAG-AAVPRNGFGDPHLHP
MLBr_00762|M.leprae_Br4923 AGPLPSNPDEDDLVALADAVRE-GPGGEHGSYGNGARASLGGFADPQLQS
Mflv_4727|M.gilvum_PYR-GCK AGPLPTHPDDDDLVALADAVREGREVPGAGVTAGFSAGLNTGFTDPV---
Mvan_1736|M.vanbaalenii_PYR-1 AGPLPSHPDDDDLVALADAVREGR---DAAPVAGFTVG----FSDPV---
MSMEG_1833|M.smegmatis_MC2_155 AGPLPTHPDEDDLVALADAVREGR--TGPGPVNGVVAG----FSDPA---
MAB_3604c|M.abscessus_ATCC_199 NGPLPSNPEDDDLVALADAVRETT---------GGVRVAAAGFSEPA---
****::*::************ . * :
Mb3286c|M.bovis_AF2122/97 AAATGAPTGGGVLAPPEPGAGRRRGHLRVLPDPAD
Rv3258c|M.tuberculosis_H37Rv AAATGAPTGGGVLAPPEPGAGRRRGHLRVLPDPAD
MMAR_1284|M.marinum_M AGAHATAPSGGVLASPEHRSGRRRGHLRVLPDPSD
MUL_2603|M.ulcerans_Agy99 AGAHATAPSGGVLASPEHRSGRRRGHLRVLPDPSD
MAV_4221|M.avium_104 GGAQATAPSSSLIAPPEQRSGRRRGHLRVLPDPSD
MLBr_00762|M.leprae_Br4923 AGAHATVPSGGLLAPSELRSGRRRGHLRVLPDPSD
Mflv_4727|M.gilvum_PYR-GCK SGAHGGALMAPPARRPET-NGRRRGHLRVLPDPAE
Mvan_1736|M.vanbaalenii_PYR-1 TGAHGGALMAPPARRPET-NGRRRGHLRVLPDPTD
MSMEG_1833|M.smegmatis_MC2_155 TGTGSGAVIAPPVRQPEP-NGRRRGHLRVLPDPTD
MAB_3604c|M.abscessus_ATCC_199 LDVSAPAPNVAPRPVHPAPVGRRRGHLRVLPDPSE
. . *************::