For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDGELKKSVDKSGNCEVSGCAEVIAEVWTLLDGECSPESSARLRHHLEECPGCLQHYGIEEQIKTLVARK CGGEKAPDGLRERLKLKISQTTVIQQRAENS*
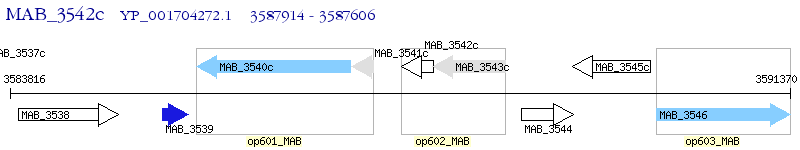
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3542c | - | - | 100% (102) | hypothetical protein MAB_3542c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3248c | - | 4e-31 | 70.13% (77) | anti-sigma factor |
| M. gilvum PYR-GCK | Mflv_4688 | - | 4e-30 | 68.83% (77) | hypothetical protein Mflv_4688 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1335 | - | 7e-32 | 72.73% (77) | anti-sigma factor |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1915 | - | 5e-30 | 69.74% (76) | anti-sigma factor, family protein |
| M. thermoresistible (build 8) | TH_0666 | - | 3e-29 | 56.38% (94) | POSSIBLE ANTI-SIGMA FACTOR |
| M. ulcerans Agy99 | MUL_2544 | - | 6e-32 | 72.73% (77) | anti-sigma factor |
| M. vanbaalenii PYR-1 | Mvan_1779 | - | 2e-29 | 67.53% (77) | hypothetical protein Mvan_1779 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4688|M.gilvum_PYR-GCK MSGIEGGGGVTD-NPEERWTP-PVGPIDPEHPECAAVIAEVWTLLDGECT
Mvan_1779|M.vanbaalenii_PYR-1 MSGTETPENETPGNSEDTWTP-PIGPIDPEHPECAAVIAEVWTLLDGECT
MSMEG_1915|M.smegmatis_MC2_155 MSETE--------REDERWTP-PIGPIDPEHPECAAVIAEVWTLLDGECT
TH_0666|M.thermoresistible__bu VSDERSG------RDDGEWSSRSVGEIDPEHPDCAAVVAEVWMLLDGECT
MMAR_1335|M.marinum_M MSEVP--------GSPDSLPD---NEDSHGYFGCAEVIAEVWTLLDGECT
MUL_2544|M.ulcerans_Agy99 MSEVP--------GSPDSLPD---NEDSHGYFGCAEVIAEVWTLLDGECT
Mb3248c|M.bovis_AF2122/97 MSENC--------GPTDAHAD---HDDSHGGMGCAEVIAEVWTLLDGECT
MAB_3542c|M.abscessus_ATCC_199 MTDGELK------KSVDKSGN-------CEVSGCAEVIAEVWTLLDGECS
:: ** *:**** ******:
Mflv_4688|M.gilvum_PYR-GCK VETRDKLRQHLEECPACLRHYGVEEKIKRLIAMKCSGERAPEGFRERLRI
Mvan_1779|M.vanbaalenii_PYR-1 VETRDKLRQHLEECPTCLRYYGVEERVKKLIAAKCSGERAPDGLRERLRI
MSMEG_1915|M.smegmatis_MC2_155 PETRDKLKQHLEECPTCLRHYGIEERVKRLIAAKCSGEKAPDSLRERLRI
TH_0666|M.thermoresistible__bu PENRDRLKRHLDECPACLRHYGVEERVKRLIAKKCSGEKAPDRLRQRLQI
MMAR_1335|M.marinum_M PETRQKLREHLEACPGCLKHYGLEERIKLLIATKCQGEKAPEGLRERVRL
MUL_2544|M.ulcerans_Agy99 PETRQKLRKHLEACPGCLKHYGLEERIKLLIATKCQGEKAPEGLRERVRL
Mb3248c|M.bovis_AF2122/97 PETRERLRRHLEACPGCLRHYGLEERIKALIGTKCRGDRAPEGLRERLRL
MAB_3542c|M.abscessus_ATCC_199 PESSARLRHHLEECPGCLQHYGIEEQIKTLVARKCGGEKAPDGLRERLKL
*. :*:.**: ** **::**:**::* *:. ** *::**: :*:*:::
Mflv_4688|M.gilvum_PYR-GCK QISQTTIIRQG----
Mvan_1779|M.vanbaalenii_PYR-1 QISQTTIIRRG----
MSMEG_1915|M.smegmatis_MC2_155 QISRTTIIRG-----
TH_0666|M.thermoresistible__bu EISRTTIIRNG----
MMAR_1335|M.marinum_M QISRTTIIRGGE---
MUL_2544|M.ulcerans_Agy99 QISRTTIIRGGE---
Mb3248c|M.bovis_AF2122/97 EIRRTTIIRGGP---
MAB_3542c|M.abscessus_ATCC_199 KISQTTVIQQRAENS
:* :**:*: