For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHIEHTFAQLGVRKEIARALREAGIEHPFAIQELTLPLALAGSDLIGQARTGMGKTYAFGVPLLHRIAT GVEDRPLNGTPRALVVVPTRELCIQVYEDLTKASKYLSAGDRDFTVVSIYGGRPYEPQIEALRAGADVVV GTPGRLLDLAQQGHLQLGGLAMLVLDEADEMLDLGFLPDIERILALAPSADAGRQSMLFSATMPDPIITL ARTFMNRPTHIRAEDPQASSVHDSTEQFVYRAHALDKIELVSRILQADGRGATMIFTRTKRTAQKVSDEL AERGFAVGAVHGDLGQIAREKALTSFRSGQINVLVATDVAARGIDIDDVTHVINYQCPEDDKTYVHRIGR TGRAGRTGIAVTLVDWDDIARWQLIDKALGLGVPEPDETYSTSPHLFTELNIPADVTGSIGEKFSGGPKR REPRGERAEGEKPKRTRTRRRTRAGQPTDESAAEAPAPEAATADGDQQAPARRRRRRRRPNATAATTASA
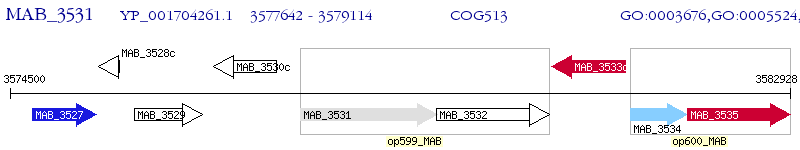
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3531 | - | - | 100% (490) | ATP-dependent DEAD-box RNA helicase |
| M. abscessus ATCC 19977 | MAB_1403 | - | 1e-81 | 39.45% (474) | cold-shock DEAD box protein A homolog (ATP-dependent RNA |
| M. abscessus ATCC 19977 | MAB_2158c | - | 1e-68 | 37.58% (463) | ATP-dependent RNA helicase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3237 | rhlE | 0.0 | 69.02% (510) | ATP-dependent RNA helicase RhlE |
| M. gilvum PYR-GCK | Mflv_4676 | - | 0.0 | 73.35% (499) | DEAD/DEAH box helicase domain-containing protein |
| M. tuberculosis H37Rv | Rv3211 | rhlE | 0.0 | 69.22% (510) | ATP-dependent RNA helicase RhlE |
| M. leprae Br4923 | MLBr_00811 | rhlE | 0.0 | 66.86% (507) | putative ATP-dependent RNA helicase |
| M. marinum M | MMAR_1346 | rhlE | 0.0 | 70.69% (505) | ATP-dependent RNA helicase RhlE |
| M. avium 104 | MAV_1401 | - | 2e-74 | 36.23% (472) | ATP-dependent rna helicase, dead/deah box family protein |
| M. smegmatis MC2 155 | MSMEG_1930 | - | 0.0 | 71.29% (505) | DEAD/DEAH box helicase |
| M. thermoresistible (build 8) | TH_1004 | rhlE | 1e-175 | 81.77% (373) | PROBABLE ATP-DEPENDENT RNA HELICASE RHLE |
| M. ulcerans Agy99 | MUL_2533 | rhlE | 0.0 | 70.50% (505) | ATP-dependent RNA helicase RhlE |
| M. vanbaalenii PYR-1 | Mvan_1791 | - | 0.0 | 73.54% (495) | DEAD/DEAH box helicase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3237|M.bovis_AF2122/97 ----------------------MTAVKHTTEST---FAKLGARDEIVRAL
Rv3211|M.tuberculosis_H37Rv ----------------------MTAVKHTTEST---FAKLGVRDEIVRAL
MMAR_1346|M.marinum_M ----------------------MTALKSTTETTPKTFAELGVRDEIVRAL
MUL_2533|M.ulcerans_Agy99 ----------------------MTALKSTTETTPKTFAELGVRDEIVRAL
MLBr_00811|M.leprae_Br4923 MNRDSKTPTQNRFSHRKAFHPHMTAPTSTAEPA---FAKLGVRDEIVRAL
Mflv_4676|M.gilvum_PYR-GCK ----------------------MTPVTTHPNPT---FAQLGVRDEIVRAL
Mvan_1791|M.vanbaalenii_PYR-1 ----------------------MTPVTTHPSPT---FAQLGVRDEIVRAL
MSMEG_1930|M.smegmatis_MC2_155 --------------------------MTQLNHS---FAELGVRDEIVRAL
MAB_3531|M.abscessus_ATCC_1997 --------------------------MSHIEHT---FAQLGVRKEIARAL
TH_1004|M.thermoresistible__bu ----------------------------------------------VRAL
MAV_1401|M.avium_104 ----------------------MTLPDSSTEAASPTFADLQIHPSVLRAI
**:
Mb3237|M.bovis_AF2122/97 GEEGIKRPFAIQELTLPLALDGEDVIGQARTGMGKTFAFGVPLLQRITSG
Rv3211|M.tuberculosis_H37Rv GEEGIKRPFAIQELTLPLALDGEDVIGQARTGMGKTFAFGVPLLQRITSG
MMAR_1346|M.marinum_M GERDIECPFAIQELTLPLALAGDDLIGQARTGMGKTFAFGVPLLQRVSSG
MUL_2533|M.ulcerans_Agy99 GERDIEGPFAIQELTLPLALAGDDLIGQARTGMGKTFAFGVPLLQRVSSG
MLBr_00811|M.leprae_Br4923 AERGIQQPFAIQELTLPLALAGDDVIGQARTGMGKTFAFGVPLLQRISVT
Mflv_4676|M.gilvum_PYR-GCK AENGIEHAFAIQELTLPLALAGDDLIGQARTGMGKTFAFGVPLLHRITTD
Mvan_1791|M.vanbaalenii_PYR-1 AENGIEHAFAIQELTLPLALAGDDLIGQARTGMGKTFAFGVPLLHRITTD
MSMEG_1930|M.smegmatis_MC2_155 AENGIEHPFAIQELTLPLALAGDDLIGQARTGMGKTYAFGVPLLHRVSSD
MAB_3531|M.abscessus_ATCC_1997 REAGIEHPFAIQELTLPLALAGSDLIGQARTGMGKTYAFGVPLLHRIATG
TH_1004|M.thermoresistible__bu AEHGIEHPFAIQELTLPLALAGEDLIGQARTGMGKTFAFGVPLLQRITTD
MAV_1401|M.avium_104 ADVGYETPTGIQAATIPALMAGSDVVGLAQTGTGKTAAFAIPILSKIDVA
: . : . .** *:* : *.*::* *:** *** **.:*:* ::
Mb3237|M.bovis_AF2122/97 DGTRPLTGAPRALVVVPTRELCLQVTDDLATAGKYLTAGPDTDDAAAVRR
Rv3211|M.tuberculosis_H37Rv DGTRPLTGAPRALVVVPTRELCLQVTDDLATAGKYLTAGPDTDDAAAVRR
MMAR_1346|M.marinum_M TGVRPLTGAPRALVVVPTRELCLQVTEDLAVAAKYLTATTETGD----QR
MUL_2533|M.ulcerans_Agy99 TGVRPLTGAPRALVVVPTRELCLQVTEDLAVAAKYLTATTETGD----QR
MLBr_00811|M.leprae_Br4923 TTARPLSGAPRGLVVVPTRELCLQVADDLATVAKYLTADDD-------NR
Mflv_4676|M.gilvum_PYR-GCK TE-RPLTGIPRALVVVPTRELCIQVYEDLVHASKYLMADDT--------R
Mvan_1791|M.vanbaalenii_PYR-1 TE-RPLTGIPRALIVVPTRELCIQVYEDLVGASKYLMADET--------R
MSMEG_1930|M.smegmatis_MC2_155 EN-RPLTGAPRALVVVPTRELCLQVYNDLAGAAKHLPTNDG--------R
MAB_3531|M.abscessus_ATCC_1997 VEDRPLNGTPRALVVVPTRELCIQVYEDLTKASKYLSAGD---------R
TH_1004|M.thermoresistible__bu TE-RPLTGIPRALVVVPTRELCLQVHDDLATAAKYLKAGD---------R
MAV_1401|M.avium_104 ST------ATQALVLAPTRELALQVAEAFSRYGAHLPKIN----------
.:.*::.*****.:** : : . :*
Mb3237|M.bovis_AF2122/97 RLSVVSIYGGRPYEPQIEALRAGADVVVGTPGRLLDLCQQGHLQLGGLSV
Rv3211|M.tuberculosis_H37Rv RLSVVSIYGGRPYEPQIEALRAGADVVVGTPGRLLDLCQQGHLQLGGLSV
MMAR_1346|M.marinum_M PLSVVSIYGGRAYEPQIEALRAGADVVVGTPGRLLDLSQQGHLQLGGLSV
MUL_2533|M.ulcerans_Agy99 PLSVVSIYGGRAYEPQIEALRAGADVVVGTPGRLLDLSQQGHLQLGGLYV
MLBr_00811|M.leprae_Br4923 RFSVVSIYGGRAYEPQIEALQTGADVVVSTPGRLLDLCQQGHLQLSGLSV
Mflv_4676|M.gilvum_PYR-GCK KLTVTAIYGGRPYEPQIEALQKGVDVVVGTPGRLLDLAQQGHLQLGGLNV
Mvan_1791|M.vanbaalenii_PYR-1 KLTVTAIYGGRPYEPQIEALQKGVDVVVGTPGRLLDLAQQGHLQLGGLSV
MSMEG_1930|M.smegmatis_MC2_155 KFTVTSIYGGRPYEPQIEALRKGVDVVVGTPGRLLDLAQQGHLQLGGLSV
MAB_3531|M.abscessus_ATCC_1997 DFTVVSIYGGRPYEPQIEALRAGADVVVGTPGRLLDLAQQGHLQLGGLAM
TH_1004|M.thermoresistible__bu KLSVVSIYGGRPYEPQIEALQKGADVVVGTPGRLLDLAQQNHLQLGGLSV
MAV_1401|M.avium_104 ---VLPIYGGSSYAVQLAGLKRGAHVVVGTPGRVIDHLERGTLDLSHVDY
* .**** .* *: .*: *..***.****::* ::. *:*. :
Mb3237|M.bovis_AF2122/97 LVLDEADEMLDLGFLPDIERILRQIP---ADRQSMLFSATMPDPIITLAR
Rv3211|M.tuberculosis_H37Rv LVLDEADEMLDLGFLPDIERILRQIP---ADRQSMLFSATMPDPIITLAR
MMAR_1346|M.marinum_M LVLDEADEMLDLGFLPDIERILRQIP---ADRQSMLFSATMPDPIITLAR
MUL_2533|M.ulcerans_Agy99 LVLDEADEMLDLGFLPDIERILRQIP---ADRQSMLFSATMPDPIITLAR
MLBr_00811|M.leprae_Br4923 LVLDEADEMLDLGFFPDIERLLGQIP---TDRQSMLFSATMPDPIITLAR
Mflv_4676|M.gilvum_PYR-GCK LVLDEADEMLDLGFLPDIERILKQIP---AKRQAMLFSATMPDPIITLAR
Mvan_1791|M.vanbaalenii_PYR-1 LVLDEADEMLDLGFLPDIERILKQIP---AKRQAMLFSATMPDPIITLAR
MSMEG_1930|M.smegmatis_MC2_155 LVLDEADEMLDLGFLPDIERILQLTP---DSRQAMLFSATMPDPIITLAR
MAB_3531|M.abscessus_ATCC_1997 LVLDEADEMLDLGFLPDIERILALAPSADAGRQSMLFSATMPDPIITLAR
TH_1004|M.thermoresistible__bu LVLDEADEMLDLGFLPDIERILRQIP---EKRQAMLFSATMPDPIITLAR
MAV_1401|M.avium_104 LVLDEADEMLTMGFAEEVDRILSETP---EYKQVALFSATMPPAIRKLTA
********** :** :::*:* * :* ******* .* .*:
Mb3237|M.bovis_AF2122/97 TFMVRPTHIRAEAPHSSAVHDATEQFVYRAHALDKVELVSRVLQARDRGA
Rv3211|M.tuberculosis_H37Rv TFMVRPTHIRAEAPHSSAVHDATEQFVYRAHALDKVELVSRVLQARDRGA
MMAR_1346|M.marinum_M TFMIQPTHIRAEAPHSLAVHDTTEQFVYRAHALDKVELVTRVLQARERGA
MUL_2533|M.ulcerans_Agy99 TFMIQPTHIRAEAPHSLAVHDTTEQFVYRAHALDKVELVTRVLQARERGA
MLBr_00811|M.leprae_Br4923 TFMDQPIHIRAEAPHSLAVHDTTQQFVYRAHALDKVELISRILQARGRGA
Mflv_4676|M.gilvum_PYR-GCK TFMTQPTHIRAEGVQGSATHDTTEQFAYRAHALDKVEMVARILQAEGRGA
Mvan_1791|M.vanbaalenii_PYR-1 TFMTQPTHIRAEAPHSSATHDSTEQFAYRAHALDKVEMVARILQAEGRGA
MSMEG_1930|M.smegmatis_MC2_155 TFMNQPTHIRAEAPHSAATHDTTAQYAYRAHALDKVELVSRILQARGRGA
MAB_3531|M.abscessus_ATCC_1997 TFMNRPTHIRAEDPQASSVHDSTEQFVYRAHALDKIELVSRILQADGRGA
TH_1004|M.thermoresistible__bu TFMNQPTHIRAEAPQSSAVHDKTKQFVYRAHALDKVEMISRILQARGRGA
MAV_1401|M.avium_104 KYLHDPLEVSTKAKTTTAEN--ISQRYIQVAGPRKMDALTRVLEVEPFEA
.:: * .: :: : : * :. . *:: ::*:*:. *
Mb3237|M.bovis_AF2122/97 TMIFTRTKRTAQKVADELTERGFAVGAVHGDLGQLAREKALKAFRTGGID
Rv3211|M.tuberculosis_H37Rv TMIFTRTKRTAQKVADELTERGFAVGAVHGDLGQLAREKALKAFRTGGID
MMAR_1346|M.marinum_M TMIFTRTKRTAQKVADELNERGFAVGAVHGDLGQVAREKALKSFRSGSVN
MUL_2533|M.ulcerans_Agy99 TMIFTRTKRTAQKVADELNERGFAVGAVHGDLGQVAREKALKSFRRGSVN
MLBr_00811|M.leprae_Br4923 TMIFTRTKRTAQKVTNELNERGFAVGAVHGDLGQIARENALEAFRTGNVD
Mflv_4676|M.gilvum_PYR-GCK TMIFTRTKRTAQKVADELGERGFRVGAVHGDLGQSAREKALKGFRNGDVD
Mvan_1791|M.vanbaalenii_PYR-1 TMIFTRTKRTAQKVADELGERGFKVGAVHGDLGQAAREKALKSFRTGEVD
MSMEG_1930|M.smegmatis_MC2_155 TMVFTRTKRTAQKVSDELAERGFKVGAVHGDLGQGAREKALKSFRTGEID
MAB_3531|M.abscessus_ATCC_1997 TMIFTRTKRTAQKVSDELAERGFAVGAVHGDLGQIAREKALTSFRSGQIN
TH_1004|M.thermoresistible__bu TMIFTRTKRTAQKVADELAERGFAVGAVHGDLGQVAREKALKAFRAGEID
MAV_1401|M.avium_104 MIVFVRTKQATEEVAERLRARGFSAAAINGDIPQGQRERTVAALKDGGID
::*.***:::::*::.* *** ..*::**: * **.:: .:: * ::
Mb3237|M.bovis_AF2122/97 VLVATDVAARGIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVT
Rv3211|M.tuberculosis_H37Rv VLVATDVAARGIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVT
MMAR_1346|M.marinum_M VLVATDVAARGIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVT
MUL_2533|M.ulcerans_Agy99 VLVATDVAARGIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVT
MLBr_00811|M.leprae_Br4923 VLVATDVAARGIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVT
Mflv_4676|M.gilvum_PYR-GCK VLVATDVAARGIDIDDITHVINFQIPEDEQAYVHRIGRTGRAGKTGIAVT
Mvan_1791|M.vanbaalenii_PYR-1 VLVATDVAARGIDIDDITHVINFQIPEDEQAYVHRIGRTGRAGKTGIAVT
MSMEG_1930|M.smegmatis_MC2_155 VLVATDVAARGIDIDDVTHVINYQCPEDEQAYVHRIGRTGRAGKTGVAIT
MAB_3531|M.abscessus_ATCC_1997 VLVATDVAARGIDIDDVTHVINYQCPEDDKTYVHRIGRTGRAGRTGIAVT
TH_1004|M.thermoresistible__bu VLVATDVAARGIDIEDVTHVINYQIPEDEQAYVHRIGRTGRAGRTGVAIT
MAV_1401|M.avium_104 ILVATDVAARGLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGTALL
:**********:*:: ::**:*:: *.* : ************::* *:
Mb3237|M.bovis_AF2122/97 LVDWDELPRWSMIDQALGLGSPDPAETYSNS-------------------
Rv3211|M.tuberculosis_H37Rv LVDWDELPRWSMIDQALGLGSPDPAETYSNS-------------------
MMAR_1346|M.marinum_M LVDWDELPRWTMIDKALGLGSPDPAETYSSS-------------------
MUL_2533|M.ulcerans_Agy99 LVDWDELPRWTMIDKALGLGSPDPAETYSSS-------------------
MLBr_00811|M.leprae_Br4923 LVDWDELHRWETIDKALGLGTPDPAETYSNS-------------------
Mflv_4676|M.gilvum_PYR-GCK LVDWDELPRWSMIDKALGLDTPDPAETYSNS-------------------
Mvan_1791|M.vanbaalenii_PYR-1 LVDWDELPRWSMIDKALGLDTPDPAETYSNS-------------------
MSMEG_1930|M.smegmatis_MC2_155 LVDWDELARWEMIDKALNLGNPDPAETYSSS-------------------
MAB_3531|M.abscessus_ATCC_1997 LVDWDDIARWQLIDKALGLGVPEPDETYSTS-------------------
TH_1004|M.thermoresistible__bu LVDWDELPRWEMIDKALNLGNPDPPETYSXXX------------------
MAV_1401|M.avium_104 FVSPRERHLLKAIEKATRQPLTEAELPTVEDVNAQRVAKFADSITAALGA
:*. : *::* .:. .
Mb3237|M.bovis_AF2122/97 --PHLYAELAIPATAGGTVGPARKSQGRR--RDT----DCDGQ----KTA
Rv3211|M.tuberculosis_H37Rv --PHLYAELAIPATAGGTVGPARKSQGRR--RDT----DCDGQ----KTA
MMAR_1346|M.marinum_M --PHLYTELSIPTDAAGTVGPPSKSGSKP--RTTEKQDDQQGE----KSG
MUL_2533|M.ulcerans_Agy99 --PHLYTELSIPTDAAGTVGPPSKSGSKP--RTTEKQDDQQGE----KSG
MLBr_00811|M.leprae_Br4923 --PHLYTELAIPAEASGTVGTPRKAQVKR--RKTDRDGDKSGTSAQAAKA
Mflv_4676|M.gilvum_PYR-GCK --PHLYDELNIPTEAGGSIGKPARSADKP--KREPREPRAERP-------
Mvan_1791|M.vanbaalenii_PYR-1 --PHLYEELNIPAEAGGSIGRPKRTADKE--PREPRE-RREKP-------
MSMEG_1930|M.smegmatis_MC2_155 --PHLYEDLDIPTDATGTVGKPAKRATKASDEDEHRISSADRP-------
MAB_3531|M.abscessus_ATCC_1997 --PHLFTELNIPADVTGSIGEKFSGGPKR---REPRGERAEGE-------
TH_1004|M.thermoresistible__bu --PSTTGAADSRAGGTWAAASAPAAATAAGSGNALVTIDAASP---ISVT
MAV_1401|M.avium_104 PGIDLFRKLVQDYEREHDVPMADIAAALAVQSRDGEEFLMAPEPPRERRE
Mb3237|M.bovis_AF2122/97 QHARNTPRRRRTRGGKPVTGHPGTNP-ISSPIVGGDATSEPGSGTASDSG
Rv3211|M.tuberculosis_H37Rv QHARNTPRRRRTRGGKPVTGHPGTNP-ISSPIVGGDATSEPGSGTASDSG
MMAR_1346|M.marinum_M RARSDAGRRRRTRGGKPVTGHPATTP-VAS----GETATETGSGSGASS-
MUL_2533|M.ulcerans_Agy99 RARSDAGRRRRTRGGKPVTGHPATTP-VAS----GETATETGSGFGASS-
MLBr_00811|M.leprae_Br4923 ARTSNSVQRHRIRGGQPVTGHPTNNPSADSLATNGKVVAETLPNSSSGS-
Mflv_4676|M.gilvum_PYR-GCK -ARDRTR--RRTRGGQSVSGHPEGTPDAADTSSSEASDSDSAGDGAPRPA
Mvan_1791|M.vanbaalenii_PYR-1 -TSNRSR--RRTRGGKSVGGHPEGTTEQAAPEADNAG--AEAPEGAASPA
MSMEG_1930|M.smegmatis_MC2_155 -ARTRTRSRRRTRGGKPLTGHPESHPEAHPESASAAPEDTEAAPAGESEA
MAB_3531|M.abscessus_ATCC_1997 -KPKRTRTRRRTRAGQPTD---ESAAEAPAPEAATADGDQQAPAR-----
TH_1004|M.thermoresistible__bu LGGAAANCCTMSRHGTPGLGWPAAAAVNNAVGDQRATTATALPANRLILL
MAV_1401|M.avium_104 RHTERRERTEKPRSTRPLATYRIAVGKRHKIGPGAIVGAIANEGGLHRSD
* .
Mb3237|M.bovis_AF2122/97 SDVVSG-------SRSGNGEAARRRRRRRRRPTHAQDG-----FAARAN-
Rv3211|M.tuberculosis_H37Rv SDVVSG-------SRSGNGEAARRRRRRRRRPTHAQDG-----FAARAN-
MMAR_1346|M.marinum_M --------------PSGN-AGATRRRRRRRKPAGVQDN-----AAASAN-
MUL_2533|M.ulcerans_Agy99 --------------PSGN-AGATRRRRRRRKPAGVQDN-----AAASAN-
MLBr_00811|M.leprae_Br4923 --------------PSTN----VRRRRHRRKPTEVAENGLDKNLAVQAN-
Mflv_4676|M.gilvum_PYR-GCK ------------------------RRRRRRRPNKATAA------GTSAGT
Mvan_1791|M.vanbaalenii_PYR-1 ------------------------RRRRRRRPSKAAAA------GSN---
MSMEG_1930|M.smegmatis_MC2_155 G-------------------EQTTRRRRRRRPRKTEAA------TAG---
MAB_3531|M.abscessus_ATCC_1997 ------------------------RRRRRRRPNATAAT------TASA--
TH_1004|M.thermoresistible__bu P----------------------IAFRRSIVPLATARK------------
MAV_1401|M.avium_104 FGHIAIGPDFSLVELPAKLPKSTLKRLEQTRISGVLINLQPDRAAAKARG
. .
Mb3237|M.bovis_AF2122/97 ------------
Rv3211|M.tuberculosis_H37Rv ------------
MMAR_1346|M.marinum_M ------------
MUL_2533|M.ulcerans_Agy99 ------------
MLBr_00811|M.leprae_Br4923 ------------
Mflv_4676|M.gilvum_PYR-GCK SAGTN-------
Mvan_1791|M.vanbaalenii_PYR-1 ------------
MSMEG_1930|M.smegmatis_MC2_155 ------------
MAB_3531|M.abscessus_ATCC_1997 ------------
TH_1004|M.thermoresistible__bu ------------
MAV_1401|M.avium_104 RDGGKPRRKYGG