For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEDARIAGTTALVQDLAWQLRGACRNANHDLFYANDDERPIERRLREQHARDVCRSCAVATRCLAYALEI EEPHGMWGGMSEAERHRIRLRVTARSSIGQ
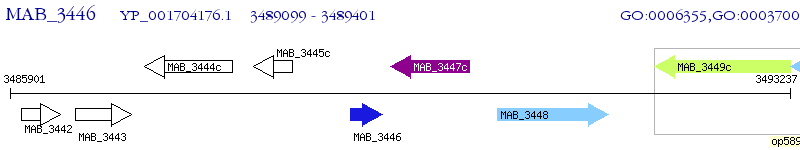
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3446 | - | - | 100% (100) | WhiB family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3726 | - | 2e-17 | 39.78% (93) | WhiB family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4343c | - | 2e-14 | 39.13% (92) | WhiB family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3450 | whiB3 | 6e-17 | 48.53% (68) | transcriptional regulatory protein WHIB-like WHIB3 |
| M. gilvum PYR-GCK | Mflv_4914 | - | 4e-16 | 37.63% (93) | transcription factor WhiB |
| M. tuberculosis H37Rv | Rv3416 | whiB3 | 6e-17 | 48.53% (68) | transcriptional regulatory protein WHIB-like WHIB3 |
| M. leprae Br4923 | MLBr_00382 | whiB3 | 9e-18 | 45.57% (79) | putative transcriptional regulator |
| M. marinum M | MMAR_1132 | whiB3 | 1e-16 | 47.06% (68) | WhiB-like regulatory protein, WhiB3 |
| M. avium 104 | MAV_4361 | - | 3e-15 | 46.97% (66) | transcription factor WhiB |
| M. smegmatis MC2 155 | MSMEG_1597 | - | 1e-17 | 40.86% (93) | transcription factor WhiB |
| M. thermoresistible (build 8) | TH_1682 | - | 2e-15 | 40.00% (80) | TRANSCRIPTIONAL REGULATORY PROTEIN WHIB-LIKE WHIB3 |
| M. ulcerans Agy99 | MUL_0895 | whiB3 | 1e-17 | 48.53% (68) | Whib-like regulatory protein, WhiB3 |
| M. vanbaalenii PYR-1 | Mvan_1506 | - | 3e-17 | 39.78% (93) | transcription factor WhiB |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3450|M.bovis_AF2122/97 MPQPEQLPGPNADIWN--WQLQGLCRGMDSSMFFHPDGERGRARTQREQR
Rv3416|M.tuberculosis_H37Rv MPQPEQLPGPNADIWN--WQLQGLCRGMDSSMFFHPDGERGRARTQREQR
MMAR_1132|M.marinum_M MPQPEQLPGPNADIWN--WQLQGLCRGVDSSMFFHPDGERGRARMQREQR
MUL_0895|M.ulcerans_Agy99 MPQPEQLPGPNADIWN--WQLQGLCRGVDSSMFFHPDGERGRARMQREQR
MAV_4361|M.avium_104 --------------------MHGLCRGVDSSMFFHPDGERGRARMQREQR
MLBr_00382|M.leprae_Br4923 MPQPKQLPGPNATIWN--WQLQGLCRGVDSSMFFHPDGERGRARMQREQR
Mflv_4914|M.gilvum_PYR-GCK MPQPQQLPGPNADIWD--WQMQGVCRGVDSAMFFHPDGERGRARAQREMR
Mvan_1506|M.vanbaalenii_PYR-1 MPQPQQLPGPNADIWD--WQIQGVCRGVDSSMFFHPDGERGRARAQREMR
MSMEG_1597|M.smegmatis_MC2_155 MPQPQQLPGPNADIWD--WQMRGLCRGVDSSMFFHPDGERGRARAQREMR
TH_1682|M.thermoresistible__bu MPQPQQLPGPNADMWD--WQMKGVCRGVDSSVFFHPDGERGRARAEREMR
MAB_3446|M.abscessus_ATCC_1997 -MEDARIAGTTALVQDLAWQLRGACRNANHDLFYANDDERPIERRLREQH
::* **. : :*: *.** * ** :
Mb3450|M.bovis_AF2122/97 AKEMCRRCPVIEACRSHALEVGEPYGVWGGLSESERDLLLKGTMGRTRGI
Rv3416|M.tuberculosis_H37Rv AKEMCRRCPVIEACRSHALEVGEPYGVWGGLSESERDLLLKGTMGRTRGI
MMAR_1132|M.marinum_M AKEMCRRCPVIEECRSHALDVGEPYGVWGGLSESERDLLLKGGIGRTRGI
MUL_0895|M.ulcerans_Agy99 AKEMCRRCPVIEECRSHALDVGEPHGVWGGLSESERDLLLKGGIGRTRGI
MAV_4361|M.avium_104 AKEMCRQCPVIQECRSHALEVGEPYGVWGGLSESERDLLLKGEIGRGRGI
MLBr_00382|M.leprae_Br4923 AKEMCRRCPVIEECRAHALDVGEPYGVWGGLSESERDLLLKGDLARSRSI
Mflv_4914|M.gilvum_PYR-GCK AKEMCRRCPVIAQCRSHALAVGEPYGIWGGLSESERELLLKRG------I
Mvan_1506|M.vanbaalenii_PYR-1 AKEMCRRCPVITQCRSHALAVGEPYGIWGGLSESERELLLKRG------I
MSMEG_1597|M.smegmatis_MC2_155 AKEMCRSCPVIAQCRSHALAVGEPYGIWGGLSESERELLLKRG------I
TH_1682|M.thermoresistible__bu AKALCRQCPVIVQCRTHALSVGEPYGIWGGLGESEREMLLKRGG-----F
MAB_3446|M.abscessus_ATCC_1997 ARDVCRSCAVATRCLAYALEIEEPHGMWGGMSEAERHRIRLRVT-----A
*: :** *.* * ::** : **:*:***:.*:**. :
Mb3450|M.bovis_AF2122/97 RRTA--
Rv3416|M.tuberculosis_H37Rv RRTA--
MMAR_1132|M.marinum_M RRTA--
MUL_0895|M.ulcerans_Agy99 RRTA--
MAV_4361|M.avium_104 RRSA--
MLBr_00382|M.leprae_Br4923 PRSA--
Mflv_4914|M.gilvum_PYR-GCK RRTA--
Mvan_1506|M.vanbaalenii_PYR-1 RRSA--
MSMEG_1597|M.smegmatis_MC2_155 RRSA--
TH_1682|M.thermoresistible__bu RRSA--
MAB_3446|M.abscessus_ATCC_1997 RSSIGQ
: