For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGAVSGALLVGGVSSDAGKSLVVTGLCRLLARKGIRVAPFKAQNMSNNSVVTVDGGEIGRAQALQARACG LEPSVRFNPVLLKPGSDRTSQLVVRGRATGNVSARSYVEHREELRQVVAEDLRTLREEYDVVICEGAGSV AEINLRTTDIANMGLARAAGLPVVLVGDIDRGGVLAHLFGSVAVLEPADQRLVAGFIVNKFRGDPTLLEP GLEQLATLTGRPTYGVLPYDDQLWLDAEDSVSVVASGQVGQTSDPIGQEWLRVAAVRLPRISNSTDIEAL ACEPGVTVRWAAHPADIADADVVVIPGTKATVADLAWMRRHGVADAVVAHARSGGPVLGICGGFQMLATT IDDRVESGAGAVEGLGLLDIDIAFEPVKTLRRWGPPGLTGYEIHHGTVTRSAVPPWLTDHDEGAVDGAVW GTHWHGVLDNDDFRRAWLTQAAAAAGRSGFAVAADTNVGDRRAAQLDRLADLVGTHVDVDALESVWEESG LPASAPSYPVIAARVE*
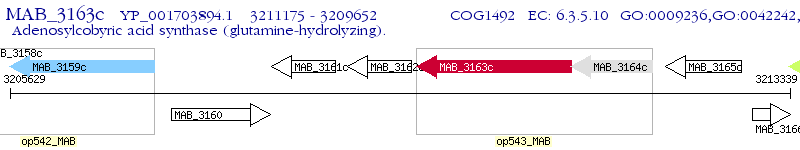
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3163c | - | - | 100% (507) | cobyric acid synthase |
| M. abscessus ATCC 19977 | MAB_0323c | - | 2e-05 | 31.51% (146) | putative cobyric acid synthase CobQ2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0261c | cobQ1 | 1e-121 | 49.80% (502) | cobyric acid synthase |
| M. gilvum PYR-GCK | Mflv_0360 | - | 1e-129 | 51.68% (505) | cobyric acid synthase |
| M. tuberculosis H37Rv | Rv0255c | cobQ1 | 1e-121 | 49.80% (502) | cobyric acid synthase |
| M. leprae Br4923 | MLBr_02327 | - | 2e-06 | 32.28% (127) | hypothetical protein MLBr_02327 |
| M. marinum M | MMAR_3838 | cobQ1 | 1e-120 | 48.89% (497) | cobyric acid synthase CobQ1 |
| M. avium 104 | MAV_3720 | cobQ | 0.0 | 69.64% (504) | cobyric acid synthase |
| M. smegmatis MC2 155 | MSMEG_2588 | cobQ | 0.0 | 66.60% (497) | cobyric acid synthase |
| M. thermoresistible (build 8) | TH_1392 | cobQ | 0.0 | 69.57% (493) | cobyric acid synthase CobQ |
| M. ulcerans Agy99 | MUL_3770 | cobQ1 | 1e-120 | 48.89% (497) | cobyric acid synthase |
| M. vanbaalenii PYR-1 | Mvan_0377 | - | 1e-127 | 50.60% (504) | cobyric acid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2588|M.smegmatis_MC2_155 ---MTGGALLVAGTTSDAGKSMLVGGLCRLLVRKGLSVAPFKAQNMSNNS
TH_1392|M.thermoresistible__bu ---MTGGALLVAGTTSDAGKSVLVAGLCRLLARKGIRVAPFKAQNMSNNS
MAV_3720|M.avium_104 ---MSG-ALLVAGTSSDAGKSVVVAGLCRLLARRGVRVAPFKAQNMSNNS
MAB_3163c|M.abscessus_ATCC_199 MTGAVSGALLVGGVSSDAGKSLVVTGLCRLLARKGIRVAPFKAQNMSNNS
MMAR_3838|M.marinum_M ---MPG--LLVAGTTSDAGKSTLTAGLCRAFARRGVRVAPFKAQNMSNNS
MUL_3770|M.ulcerans_Agy99 ---MPG--LLVAGTTSDAGKSTLTAGLCRAFARRGVRVAPFKAQNMSNNS
Mb0261c|M.bovis_AF2122/97 ---MSG--LLVAGTTSDAGKSAVTAGLCRALARRGVRVAPFKAQNMSNNS
Rv0255c|M.tuberculosis_H37Rv ---MSG--LLVAGTTSDAGKSAVTAGLCRALARRGVRVAPFKAQNMSNNS
Mflv_0360|M.gilvum_PYR-GCK ---MTG--LLVAGTTSDAGKTAVTTGLCRALARRGVKVAPYKAQNMSNNS
Mvan_0377|M.vanbaalenii_PYR-1 ---MTG--LLIAGTTSDAGKTAVTTGLCRALARRGLKVAPYKAQNMSNNS
MLBr_02327|M.leprae_Br4923 --------------------------------------------------
MSMEG_2588|M.smegmatis_MC2_155 AVT----VEGGEIGRAQAMQARAAGLAPSVRFNPILLKPGGDRTSQLVVR
TH_1392|M.thermoresistible__bu AVT----VEGGEIGRAQAMQARAAGLEPSVRFNPILLKPGGDRTSQLVVR
MAV_3720|M.avium_104 AVT----VEGGEIGRAQAIQARAAGLEPSVRFNPVLLKPGSDRTSQLVIK
MAB_3163c|M.abscessus_ATCC_199 VVT----VDGGEIGRAQALQARACGLEPSVRFNPVLLKPGSDRTSQLVVR
MMAR_3838|M.marinum_M MVCQGPGGAGVEIGRAQWVQALAANATPEAAMNPVLLKPGSDHRSHVVLM
MUL_3770|M.ulcerans_Agy99 MVCQGPGGAGVEIGRAQWVQALAANATPEAAMNPVLLKPGSDHRSHVVLM
Mb0261c|M.bovis_AF2122/97 MVCRGPDGTGVEIGRAQWVQALAARTTPEAAMNPVLLKPASDHRSHVVLM
Rv0255c|M.tuberculosis_H37Rv MVCRGPDGTGVEIGRAQWVQALAARTTPEAAMNPVLLKPASDHRSHVVLM
Mflv_0360|M.gilvum_PYR-GCK MVCTSVDGAGAEIGRAQWVQALAARATPEPAMNPVLLKPGSDRRSHVVLM
Mvan_0377|M.vanbaalenii_PYR-1 MVCAGDDG-GVEIGRAQWVQALAARATPEAAMNPVLLKPGSDRRSHVVLM
MLBr_02327|M.leprae_Br4923 --------------------------------------------------
MSMEG_2588|M.smegmatis_MC2_155 GQVTGSVAAADYINHRDHLAAVVADELSSLREDFDAVICEGAGSPAEINL
TH_1392|M.thermoresistible__bu GEVSGSVAAGDYIRHRDRLRAVVAEELTALRTEFDVVLCEGAGSPAEINL
MAV_3720|M.avium_104 GQVAESVTATSYVQHRDRLAALVLNELTCLRDEFDAVICEGAGSPAEINL
MAB_3163c|M.abscessus_ATCC_199 GRATGNVSARSYVEHREELRQVVAEDLRTLREEYDVVICEGAGSVAEINL
MMAR_3838|M.marinum_M GRSWGELASSNWFEGRQVLAETAHRAFDDLAARYDVVVAEGAGSPAEINL
MUL_3770|M.ulcerans_Agy99 GRSWGELASSNWFEGRQVLADTAHRAFDDLAARYDVVVAEGAGSPAEINL
Mb0261c|M.bovis_AF2122/97 GKPWGEVASSSWCAGRRALAEAACRAFDALAARYDVVVAEGAGSPAEINL
Rv0255c|M.tuberculosis_H37Rv GKPWGEVASSSWCAGRRALAEAACRAFDALAARYDVVVAEGAGSPAEINL
Mflv_0360|M.gilvum_PYR-GCK GRPWGQVSSSDWLEGRRALATAAHEAFDELASRYEVVVAEGAGSPTEINL
Mvan_0377|M.vanbaalenii_PYR-1 GRPWGHVSSSDWLEGRRALAAAAHAAYDDLAGRYDVIVAEGAGSPTEINL
MLBr_02327|M.leprae_Br4923 --------------------------------------------------
MSMEG_2588|M.smegmatis_MC2_155 RATDLANMGLARAAALPVIVVGDIDRGGLLAHLHGTVAVLEPADQALVSG
TH_1392|M.thermoresistible__bu RATDLANLGLARAADLPIVLIGDIDRGGLLAHFFGTVAVLEPADQRLIAG
MAV_3720|M.avium_104 RATDLANMGLARAADLAVILVGDIDRGGLLAHLFGTVAVLDPQDQALIAG
MAB_3163c|M.abscessus_ATCC_199 RTTDIANMGLARAAGLPVVLVGDIDRGGVLAHLFGSVAVLEPADQRLVAG
MMAR_3838|M.marinum_M RAGDYVNMGLACHAELPTIVVGDIDRGGVFAAFFGTIALLSAEDQALVAG
MUL_3770|M.ulcerans_Agy99 RAGDYVNMGLARHAELPTIVVGDIDRGGVFAAFFGTIALLSVEDQALVAG
Mb0261c|M.bovis_AF2122/97 RAGDYVNMGLARHAGLPTIVVGDIDRGGVFAAFLGTVALLAAEDQALVAG
Rv0255c|M.tuberculosis_H37Rv RAGDYVNMGLARHAGLPTIVVGDIDRGGVFAAFLGTVALLAAEDQALVAG
Mflv_0360|M.gilvum_PYR-GCK RAGDYVNLGLARHAGLPTVVVGDIDRGGVFAAFFGTVALLSPQDQALIAG
Mvan_0377|M.vanbaalenii_PYR-1 RAGDYVNLGLARHAGLPTVVVGDIDRGGVFAAFFGTVALLSPEDQALIAG
MLBr_02327|M.leprae_Br4923 ----------MRIGLVLPDVMGTYGDGGNAVVLRQRLLLRG------IAA
. : ::* . ** . : : : ::.
MSMEG_2588|M.smegmatis_MC2_155 FVVNKFRGDPSLLAPGLRQLAELTGRPTYGVIPFHDEIWLDTEDSVSVRP
TH_1392|M.thermoresistible__bu FVVNKFRGDPALLTPGLDELHRRTGRPTYGIIPFSDDLWLDAEDSVSVVS
MAV_3720|M.avium_104 FVVNKFRGDPALLQPGLRRLHDITGRPTYGVLPYADGLWLDAEDSLSVLA
MAB_3163c|M.abscessus_ATCC_199 FIVNKFRGDPTLLEPGLEQLATLTGRPTYGVLPYDDQLWLDAEDSVSVVA
MMAR_3838|M.marinum_M FVVNKFRGDLDLLAPGLRDLEGVTGRQVFGTLPWHADLWLDSEDALDLTG
MUL_3770|M.ulcerans_Agy99 FVVNKFRGDLDLLAPGLRDLERVTGRQVFGTLPWHADLWLDSEDALDLTG
Mb0261c|M.bovis_AF2122/97 FVVNKFRGDSDLLAPGLRDLERVTGRRVYGTLPWHPDLWLDSEDALDLQG
Rv0255c|M.tuberculosis_H37Rv FVVNKFRGDSDLLAPGLRDLERVTGRRVYGTLPWHPDLWLDSEDALDLQG
Mflv_0360|M.gilvum_PYR-GCK FVVNKFRGDVDLLAPGLRDLERLTGRRVYGTLPWHPDIWLDSEDALELAG
Mvan_0377|M.vanbaalenii_PYR-1 FVVNKFRGDPALLAPGLRDLERLTGRRVYGTLPWHPDLWLDSEDALELQG
MLBr_02327|M.leprae_Br4923 EIVELTLADP---VPESLDLYTLGGAEDY---------------------
:*: .* * * * :
MSMEG_2588|M.smegmatis_MC2_155 GGLVGAPEPPRGEQTLTVAAIRLPRISNSTDIEALACEPGVVVRWVTDAA
TH_1392|M.thermoresistible__bu GRVVGTPQPPRGEQSLTVAAVRLPRISNSTDVEALACEPGVVVRWVADPT
MAV_3720|M.avium_104 HRVVGAPDPPLGDDWLRVAAVRLPRISNSTDVEALACEPGVLVRWVSEPS
MAB_3163c|M.abscessus_ATCC_199 SGQVGQTSDPIGQEWLRVAAVRLPRISNSTDIEALACEPGVTVRWAAHPA
MMAR_3838|M.marinum_M RRAASTG-------AHRVAVVRLPRISNFTDVDALGLEPDLDVVFASDPR
MUL_3770|M.ulcerans_Agy99 RRAASAG-------AHRVAVVRLPRISNFTDVDALGLEPDLDVVFASDPR
Mb0261c|M.bovis_AF2122/97 RRAAGTG-------ARRVAVVRLPRISNFTDVDALGLEPDLDVVFASDPR
Rv0255c|M.tuberculosis_H37Rv RRAAGTG-------ARRVAVVRLPRISNFTDVDALGLEPDLDVVFASDPR
Mflv_0360|M.gilvum_PYR-GCK RRSAQSG-------ARRVAVIRLPRISNFTDVDALGLEPDLDVVFASHPG
Mvan_0377|M.vanbaalenii_PYR-1 RRSARPG-------ARRVAVVRLPRISNFTDVDAFGLEPDLDVVFASHPS
MLBr_02327|M.leprae_Br4923 ------------------------------------------------AQ
.
MSMEG_2588|M.smegmatis_MC2_155 DLTGADLVVIPGSKATVTDLRWLRERGLAAGIAAHAAAGRAVLGVCGGFQ
TH_1392|M.thermoresistible__bu ELTDADAVVLPGSKATVADLTWLRDRGLADAIVAHAAAGRAVLGVCGGFQ
MAV_3720|M.avium_104 DVADADLVVIPGSKATVADLSWVRERGLADAITAHAAAGRPVLGICGGFQ
MAB_3163c|M.abscessus_ATCC_199 DIADADVVVIPGTKATVADLAWMRRHGVADAVVAHARSGGPVLGICGGFQ
MMAR_3838|M.marinum_M GLGDADLIVVPGTRATIADLAWLRARGLDRALMAHVAAGKPLLGICGGFQ
MUL_3770|M.ulcerans_Agy99 GLGDADLIVVPGTRATIADLAWLRARGLDRALMAHVAAGKPLLGICGGFQ
Mb0261c|M.bovis_AF2122/97 ALDDADLIVLPGTRATIADLAWLRARDLDRALLVHVAAGKPLLGICGGFQ
Rv0255c|M.tuberculosis_H37Rv ALDDADLIVLPGTRATIADLAWLRARDLDRALLVHVAAGKPLLGICGGFQ
Mflv_0360|M.gilvum_PYR-GCK SLADADLVVLPGTRATIADLAWLRSRGLDSALRRHVAAGRPVLGICGGFQ
Mvan_0377|M.vanbaalenii_PYR-1 ALADADLVVLPGTRSTIADLAWLRSRGLDRAVLAHAAAGRPVLGICGGFQ
MLBr_02327|M.leprae_Br4923 RLATRHLLRYPG-------------------LQHAASYGAPVLAICAAVQ
: . : ** : . * .:*.:*...*
MSMEG_2588|M.smegmatis_MC2_155 MLCSRIDDP--VESR---EGRVDGLGLLDADIEFAAQKTLRH--------
TH_1392|M.thermoresistible__bu MLCRRIDDR--VESG---VGRVDGLGLLDADIEFAADKTLRR--------
MAV_3720|M.avium_104 MLCRRIDDR--VESG---AGEVAGLGLLDADIAFHPTKTLRR--------
MAB_3163c|M.abscessus_ATCC_199 MLATTIDDR--VESG---AGAVEGLGLLDIDIAFEPVKTLRR--------
MMAR_3838|M.marinum_M MLGRVIRDPDGVEGP---VAEADGLGLLDVETTFGAEKVLRLPRGQ----
MUL_3770|M.ulcerans_Agy99 MLGRVIRDPDGVEGP---VAEADGLGLLDVETTFGAEKVLRLPRGQ----
Mb0261c|M.bovis_AF2122/97 MLGRVIRDPYGIEGPGGQVTEVEGLGLLDVETAFSPHKVLRLPRGE----
Rv0255c|M.tuberculosis_H37Rv MLGRVIRDPYGIEGPGGQVTEVEGLGLLDVETAFSPHKVLRLPRGE----
Mflv_0360|M.gilvum_PYR-GCK MLGRVIRDPHGVEGP---VAGVDGLGLLDVETTFGPDKVLRLPSGR----
Mvan_0377|M.vanbaalenii_PYR-1 MLGRVIRDPHGVEGG---TSEADGLGLLDIETDFVADKALRLPEGQ----
MLBr_02327|M.leprae_Br4923 VLGQWYETAS--------RERVDGVGLLDVTTAPQDVRTIGELVSTPLLT
:* . *:**** :.:
MSMEG_2588|M.smegmatis_MC2_155 WETP-LHGYEIHHGQVAR-SAETDWLG-----IGLRRGAVYGTHWHGLLD
TH_1392|M.thermoresistible__bu WERP-LRGYEIHHGQVTR-SAADDWHG-----VGLRRGAVCGTHWHGLLD
MAV_3720|M.avium_104 WQRP-LAGYEIHHGRVSR-CAEHSWFDSDAEPQGLVRGAVFGTHWHGLLD
MAB_3163c|M.abscessus_ATCC_199 WGPPGLTGYEIHHGTVTR-SAVPPWLT--DHDEGAVDGAVWGTHWHGVLD
MMAR_3838|M.marinum_M GLGVTASGYEIHHGRITAGDAAQQFLG------GARDGQVFGTMWHGSLE
MUL_3770|M.ulcerans_Agy99 GLGVTASGYEIHHGRITAGDAAQQFLG------GARDGQVFGTMWHGSLE
Mb0261c|M.bovis_AF2122/97 GLGVPASGYEIHHGRITRGDTAEEFLG------GARDGPVFGTMWHGSLE
Rv0255c|M.tuberculosis_H37Rv GLGVPASGYEIHHGRITRGDTAEEFLG------GARDGPVFGTMWHGSLE
Mflv_0360|M.gilvum_PYR-GCK WFGAPATGYEIHHGRITRGDGVDEFLD------GARCGQVFGTMWHGALE
Mvan_0377|M.vanbaalenii_PYR-1 WEATSASGYEIHHGRITPGPGGDEFPG------GVRCGPVFGTMWHGAFE
MLBr_02327|M.leprae_Br4923 SLNQRLSGFENHRGGTVLGPAATPLGA-----VVKGAGNRVGDHFDGVIQ
*:* *:* * * :.* ::
MSMEG_2588|M.smegmatis_MC2_155 NDALRRDWLTEVAAAAGRDGFVVADDVDVSARRDAQLDLMADLIENHLDV
TH_1392|M.thermoresistible__bu NDELRRQWLSEAAVAAGRHGFVVADDIDVGARRDRQLDQIADLLTAHLDL
MAV_3720|M.avium_104 NDDFRRAWLVRVAEAAGRR--FVPGDTDVAARRDAQLDLAADLLAAHLDV
MAB_3163c|M.abscessus_ATCC_199 NDDFRRAWLTQAAAAAGRSGFAVAADTNVGDRRAAQLDRLADLVGTHVDV
MMAR_3838|M.marinum_M GDALREAFLRETLGLTGSG-------TSFSAARERRLDLLGDLVERHLDV
MUL_3770|M.ulcerans_Agy99 GDALREAFLRETLGLTESG-------TSFSAARERRLDLLGDLVERHLDV
Mb0261c|M.bovis_AF2122/97 GDALREAFLRETLGLAPSG-------SCFLAARERRLDLLGDLVERHLDV
Rv0255c|M.tuberculosis_H37Rv GDALREAFLRETLGLAPSG-------SCFLAARERRLDLLGDLVERHLDV
Mflv_0360|M.gilvum_PYR-GCK GDELRSRFLQEALGVAPSG-------VSFPAAREARLDLLGDLVERHLDV
Mvan_0377|M.vanbaalenii_PYR-1 GDALRARFLTETLGVPASG-------ASFPKAREDRIDLLGDLVEEHLDV
MLBr_02327|M.leprae_Br4923 G---------SVMATYMHG--------PCLARNPELADLLLSKVVGELAP
. . . * . : .:
MSMEG_2588|M.smegmatis_MC2_155 GAILDLLEH-----GAPHRPTMSTALHV
TH_1392|M.thermoresistible__bu DALFALLED-----GPPPRPVLRTDVI-
MAV_3720|M.avium_104 DAVLGLLD------GPPPRPRLATRLCR
MAB_3163c|M.abscessus_ATCC_199 DALESVWEESGLPASAPSYPVIAARVE-
MMAR_3838|M.marinum_M DALLALARHG----CAPALPFLPPGAP-
MUL_3770|M.ulcerans_Agy99 DALLALARHG----CAPALPFLPPGAP-
Mb0261c|M.bovis_AF2122/97 DALLNLARHG----CPPTLPFLAPGAP-
Rv0255c|M.tuberculosis_H37Rv DALLNLARHG----CPPTLPFLAPGAP-
Mflv_0360|M.gilvum_PYR-GCK DALLDLAKTG----PVAGLPFLPPGAP-
Mvan_0377|M.vanbaalenii_PYR-1 DALLRLAEHA----PPQGLPFLPPGSP-
MLBr_02327|M.leprae_Br4923 LELPEVELLR--------RERLAAR---
: : : .