For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQPAELAQAAMTAALMGAIAVVSIVLPGAVVFAWLGAVPMGVLCYRHRIRVALAACVAAGLISFLIAGFG GLVSALTCAYMGAISGQVRRRSRGAATMFTVAALWGVLVSTFCVGVFAALRNLREVVLGAVAANVGGFAT LLQGVPVLGRGAAGLDHAVAGWLVHWPWFFGVSVWLIVIFGTSFGWRVLTPVLRRLEEVTDLASVGTLPA PHESTAPGPLPTVLTDVGYRYAGAERDALAPVTMRVDVGEHIAVTGANGTGKSTLMRILAGVPPTTGSVE RPAGVGLGQVGGTALVLQHPESQVLGLRVGDDIVWGLPPDRDIDIEGLLTEVGLSGMAGRDTAGLSGGEL QRLAVASALAREPALLIADEVTTMVDHDGRQTLINVLDGLTTHHRLGLVHITHYPQEADAADRVVALGGI AVRPPAEPPSPAESPSKETILDVRSVSFDYAVGTPWSQPVLRDVSFRVDSGDGILLCGGNGSGKSTLAWI MAGLLDPSAGQCLLDGRPTAEQVGAVALCFQAARLQLLRGHAGAAVAALAGYSAADTDSIDRALASVSLD PAIGGVLIDRLSGGQLRRVALAGLLARSPRLLILDEPLAGLDVDAQADLIDLLIRIRNGGQAVIVISHDT DSLSPLCPRTIRLEQGELVTAG
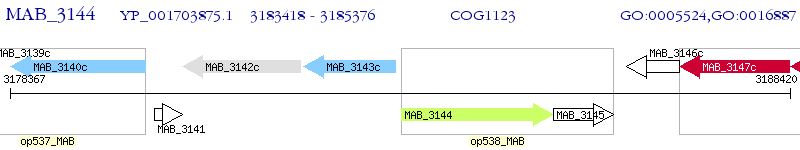
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3144 | - | - | 100% (652) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1713 | - | 0.0 | 52.81% (659) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_0429 | - | 8e-29 | 28.36% (483) | putative oligopeptide ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2353c | - | 1e-179 | 50.83% (663) | putative transmembrane ATP-binding protein ABC transorter |
| M. gilvum PYR-GCK | Mflv_2730 | - | 0.0 | 53.25% (661) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2326c | - | 1e-179 | 50.83% (663) | transmembrane ATP-binding protein ABC transorter |
| M. leprae Br4923 | MLBr_00848 | - | 1e-177 | 51.29% (657) | ABC transporter |
| M. marinum M | MMAR_3626 | - | 1e-180 | 51.82% (658) | transmembrane ATP-binding protein ABC transorter |
| M. avium 104 | MAV_2082 | - | 1e-179 | 52.48% (646) | ABC transporter |
| M. smegmatis MC2 155 | MSMEG_4470 | - | 0.0 | 54.30% (663) | ABC transporter |
| M. thermoresistible (build 8) | TH_0730 | - | 0.0 | 50.45% (664) | POSSIBLE TRANSMEMBRANE ATP-BINDING PROTEIN ABC TRANSORTER |
| M. ulcerans Agy99 | MUL_1239 | - | 0.0 | 51.82% (658) | transmembrane ATP-binding protein ABC transorter |
| M. vanbaalenii PYR-1 | Mvan_3806 | - | 0.0 | 54.53% (662) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2730|M.gilvum_PYR-GCK ----------------MSAAVRPTSPPEAGRAGQFSGDWQNREMTSGRSE
Mvan_3806|M.vanbaalenii_PYR-1 -------------------------------------------MTSAGPE
TH_0730|M.thermoresistible__bu -------------------------------------------MTMTGQA
MSMEG_4470|M.smegmatis_MC2_155 -------------------------------------------MTANRPA
Mb2353c|M.bovis_AF2122/97 ----------------------------MCCAVCGPEPGRIGEVTPLGPC
Rv2326c|M.tuberculosis_H37Rv ----------------------------MCCAVCGPEPGRIGEVTPLGPC
MMAR_3626|M.marinum_M -------------------------------------------MTASAPL
MUL_1239|M.ulcerans_Agy99 -------------------------------------------MTASAPL
MLBr_00848|M.leprae_Br4923 MDLCIHRRMALLCQATRRCHRWKTQLGRCAAPDLAPGTGTIGKVTAHDPR
MAV_2082|M.avium_104 --------------------------------------------------
MAB_3144|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2730|M.gilvum_PYR-GCK SDTRRTGRLQPVELAQAAVMAALCAAIAIIAVVVPFAAGLSLVGTVPMGL
Mvan_3806|M.vanbaalenii_PYR-1 SAPRRTGPLQPVELAQAAVMAALCAAIAIIAVVVPFAGSLSLLGTVPMGL
TH_0730|M.thermoresistible__bu RRPHRAGSLHPVELAHASVMAALTAATAIIAVVVPFAAGVSLLGTVPMGM
MSMEG_4470|M.smegmatis_MC2_155 RAPRRG--LTPGELAQASVMAALCAATAIIAVVVPFAAGLSVLGTVPMGL
Mb2353c|M.bovis_AF2122/97 PAQHRGGPLRPSELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
Rv2326c|M.tuberculosis_H37Rv PAQHRGGPLRPSELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MMAR_3626|M.marinum_M APPHRGGPLRPGELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MUL_1239|M.ulcerans_Agy99 APPHRGGPLRPGELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MLBr_00848|M.leprae_Br4923 HLHHRSGSLQPGELAQASVLAALCAVTAIVSVVVPFAAGLALLGTVPMGL
MAV_2082|M.avium_104 -------------------MGALCAAIEILAAVIPFAQGLGVLGTVPMGL
MAB_3144|M.abscessus_ATCC_1997 --------MQPAELAQAAMTAALMGAIAVVSIVLPGAVVFAWLGAVPMGV
.** .. ::: *:* * .. :*:** *:
Mflv_2730|M.gilvum_PYR-GCK LAYRHRMRVLLAATVAGGLVAFLIAGMGGLMTVINCAYIGGLTGIVKRRG
Mvan_3806|M.vanbaalenii_PYR-1 LAYRYRLRVLLAATVAGGLVAFLIAGMGGFMTVVNCAYIGGLTGIVKRRG
TH_0730|M.thermoresistible__bu LAYRFRFRVLIAATVAGATIAFLIAGMGGFMTVVNCAYIGGLTGTVKRRG
MSMEG_4470|M.smegmatis_MC2_155 LAYRYRLRVLLAATVAAGVIAFLIAGLGGMMTVFNCAYIGGLTGIVKRRG
Mb2353c|M.bovis_AF2122/97 LAYRYRLRVLAAATVAAGMIAFLIAGLGGFMGVVHSAYIGGLTGIVKRRG
Rv2326c|M.tuberculosis_H37Rv LAYRYRLRVLAAATVAAGMIAFLIAGLGGFMGVVHSAYIGGLTGIVKRRG
MMAR_3626|M.marinum_M LAYRYRLRVLVAATVAAGVIAFLIAGLGGFMGVVHSAYIGGLTGIVKRKG
MUL_1239|M.ulcerans_Agy99 LAYRYRLRVLVAATVAAGVIAFLIAGLGGFMGVVHSAYIGGLTGIVKRKG
MLBr_00848|M.leprae_Br4923 LAYRYRFRVLMTAMVAAGVIAFLITGLGGFIAVVNSAYIGGLTGVVKRKG
MAV_2082|M.avium_104 LAYRYRPRALMAATVAGGVIAFLIAGLGSLFMLVDCAWVGGLCGIVKRKG
MAB_3144|M.abscessus_ATCC_1997 LCYRHRIRVALAACVAAGLISFLIAGFGGLVSALTCAYMGAISGQVRRRS
*.**.* *. :* **.. ::***:*:*.:. . .*::*.: * *:*:.
Mflv_2730|M.gilvum_PYR-GCK RGTPTVFVCALVAGALFGIVIVAALGVLSRLRNLIFDSVTANIAGTAAVL
Mvan_3806|M.vanbaalenii_PYR-1 RGTPTVVACALVAGALFGVAVVAALALLSRLRHLIFDSMTANITGLAAIL
TH_0730|M.thermoresistible__bu RGTGTVLVAALIAGAAFGAAIVGALTVLSRLRHLMFDTITANVHGVARII
MSMEG_4470|M.smegmatis_MC2_155 RGTPTVFAAAIVAGALFGAMIVVALLVLSRLRNLMFDTLTANIDGLAAVF
Mb2353c|M.bovis_AF2122/97 RGTPTVVVSSLIGGFVFGAAMVGMLAAMVRLRHLIFKVMTANVDGIAATL
Rv2326c|M.tuberculosis_H37Rv RGTPTVVVSSLIGGFVFGAAMVGMLAAMVRLRHLIFKVMTANVDGIAATL
MMAR_3626|M.marinum_M RGTPTVIVSSLIAGLAFGSAMVGMLAALTRLRHLIFKVMTANVEGLSSFM
MUL_1239|M.ulcerans_Agy99 RGTPTVIVSSLIAGLAFGSAMVGMLAALTRLRHLIFKVMTANVEGLSSFM
MLBr_00848|M.leprae_Br4923 QGTLTVIALALFAGLAFGAANVVALVVLGRLRHLIFKAMTANVDGIAATL
MAV_2082|M.avium_104 RGTPTVALLSLIAGVLWGAGWVAVLAVLTRLRHLFFDVITANANGVAAFL
MAB_3144|M.abscessus_ATCC_1997 RGAATMFTVAALWGVLVSTFCVGVFAALRNLREVVLGAVAANVGGFATLL
:*: *: : . * . * : : .**.:.: ::** * : :
Mflv_2730|M.gilvum_PYR-GCK ERVPGLAPVGERLTRDFATMLHYWPFLFFASGVVSITFVSLVGWWALSRV
Mvan_3806|M.vanbaalenii_PYR-1 ERVPGLEPVAERLTRDFATALHYWPYLFFAVGVLSITFVSLVGWWALSRV
TH_0730|M.thermoresistible__bu SGIPNMSGAGERLRRDFETMLSYWPVLILAAGVISITVVTLIGWWALSRV
MSMEG_4470|M.smegmatis_MC2_155 ARVPVLEGAAEPLKTMFATLLHHWPVLMLLMGISSITMVTMVGWWALSRV
Mb2353c|M.bovis_AF2122/97 ARMH-MQGAAADVKRYFAEGLQYWPWVLLGYFNIGIMIVSLIGWWALSRL
Rv2326c|M.tuberculosis_H37Rv ARMH-MQGAAADVKRYFAEGLQYWPWVLLGYFNIGIMIVSLIGWWALSRL
MMAR_3626|M.marinum_M ARMG-MEGAAEDLKRYFAEGLHYWPWVLLGYFTVAIMVVSLIGWWALSRL
MUL_1239|M.ulcerans_Agy99 ARMG-MEGAAEDLKRYFAEGLHYWPWVLLGYFTVAIMVVSLIGWWALSRL
MLBr_00848|M.leprae_Br4923 TWMH-LPWVAVQLKRYFADGLQHWPWMLLGYFVITILVVSLIGWWVLSRV
MAV_2082|M.avium_104 NWMH-LQGVGAGLKRYVADGLQHWPLLIFPYMILLVVVVSFISWSALSRL
MAB_3144|M.abscessus_ATCC_1997 QGVPVLGRGAAGLDHAVAGWLVHWPWFFGVSVWLIVIFGTSFGWRVLTPV
: : . : . * :** .: : . : ..* .*: :
Mflv_2730|M.gilvum_PYR-GCK LKRLAGVPDVHKLEAS---ADTGEIAPTPLTLTDVRFRYPGTDHDALGPV
Mvan_3806|M.vanbaalenii_PYR-1 LGRLLGVPDVHKLEAS---NDVGSIAPVPLQLRDVRFRYPNAGQDALGPV
TH_0730|M.thermoresistible__bu LARLVVIPDVHKLESF---EDTGPVAPVPLRLRDVRFRYPNADRDALGPL
MSMEG_4470|M.smegmatis_MC2_155 LGRLLGVPDVHKLDSP---PEHGPIGPVPAQLTDVRFRYPGADHDALGPV
Mb2353c|M.bovis_AF2122/97 LERMRGIPDVHKLDPPPGDDVDALIGPVPVRLDKVRFRYPRAGQDALREV
Rv2326c|M.tuberculosis_H37Rv LERMRGIPDVHKLDPPPGDDVDALIGPVPVRLDKVRFRYPRAGQDALREV
MMAR_3626|M.marinum_M LERMRGIPDVHKLDAP-VDGEATSIEPVPVRLDKVRFRYPGAGQDALREV
MUL_1239|M.ulcerans_Agy99 LERMRGIPDVHKLDAP-VDGEATSIEPVPVRLDKVRFRYPGAGQDALREV
MLBr_00848|M.leprae_Br4923 LERIRDIPDVHKLDAPSACNEDAPVGPVPVWLDKVRFRYPHAGQDALREV
MAV_2082|M.avium_104 LDRMRKIPDVHKLDPP--DDGHAAIGPVPVRLENVRFRYPGAEQDALREV
MAB_3144|M.abscessus_ATCC_1997 LRRLEEVTDLASVGTLPAPHESTAPGPLPTVLTDVGYRYAGAERDALAPV
* *: :.*: .: . * * * .* :**. : :*** :
Mflv_2730|M.gilvum_PYR-GCK SLAVQPGEHVAVTGANGSGKTTLMLVLSGREPTGGTVERPGAVGLGRIGG
Mvan_3806|M.vanbaalenii_PYR-1 SLAVQPGEHLAVTGANGSGKTTLMLVLSGREATAGTVERPGAVGLGKIGG
TH_0730|M.thermoresistible__bu TMTVQPGEHVAITGANGSGKTTLMLLLAGREPTAGTVERPGAVGLGRMGG
MSMEG_4470|M.smegmatis_MC2_155 TMDLRVGEHVAVTGPNGSGKTTLMLVLAGREPTSGSVERAGAVGLGRMGG
Mb2353c|M.bovis_AF2122/97 SLDVRAGEHLAIIGANGSGKTTLMLILAGRAPTSGTVDRPGTVGLGKLGG
Rv2326c|M.tuberculosis_H37Rv SLDVRAGEHLAIIGANGSGKTTLMLILAGRAPTSGTVDRPGTVGLGKLGG
MMAR_3626|M.marinum_M SLDVRVGEHIAITGANGSGKTTLMLILAGREPTSGTVERPGAVGLGKLGG
MUL_1239|M.ulcerans_Agy99 SLDVRVGEHIAITGANGSGKTTLMLILAGREPTSGTVERPGAVGLGKLGG
MLBr_00848|M.leprae_Br4923 SLDLRVGEHVAVTGANGSGKTTLMLILAGREPTSGTVDRPGAVGLGKLGG
MAV_2082|M.avium_104 SLDIQAGEHVAVTGANGSGKTTLMLILAGRQPTSGTVHRPGAVGLGDVDG
MAB_3144|M.abscessus_ATCC_1997 TMRVDVGEHIAVTGANGTGKSTLMRILAGVPPTTGSVERPAGVGLGQVGG
:: : ***:*: *.**:**:*** :*:* .* *:*.*.. **** :.*
Mflv_2730|M.gilvum_PYR-GCK TAVVMQHPESQVLGTRVADDLVWGLPPGTAVDVDRLLGEVGLDGLAERDT
Mvan_3806|M.vanbaalenii_PYR-1 TAVVMQHPESQVLGTRVADDVVWGLPPGASIDVDRLLAEVGLDGLAERDT
TH_0730|M.thermoresistible__bu TAVVMQHPESQVLGTRVADDVVWGLPPGTATDMPRLLEEVGLDGLAERDT
MSMEG_4470|M.smegmatis_MC2_155 TAVVMQHPESQVLGTRVADDVVWGLPPGTATDVDRLLSEVGLDGMAERDT
Mb2353c|M.bovis_AF2122/97 TAVVLQHPESQVLGTRVADDVVWGLPLGTTADVGRLLSEVGLEALAERDT
Rv2326c|M.tuberculosis_H37Rv TAVVLQHPESQVLGTRVADDVVWGLPLGTTADVGRLLSEVGLEALAERDT
MMAR_3626|M.marinum_M TAVVLQHPESQVLGTRVADDVVWGLPPGTKIDVDQQLAEVGLEGLAERET
MUL_1239|M.ulcerans_Agy99 TAVVLQHPESQVLGTRVADDVVWGLPPGTKIDVDQQLAEVGLEGLAERET
MLBr_00848|M.leprae_Br4923 TAVVLQHPESQVLGTRVADDVVWGLPPGTDVDVNRLLREVGLDAFAERDT
MAV_2082|M.avium_104 TAIVLQHPKSQVLGTRVADDVVWGLPPGTDIDVHRLLREVGLDGLAERDT
MAB_3144|M.abscessus_ATCC_1997 TALVLQHPESQVLGLRVGDDIVWGLPPDRDIDIEGLLTEVGLSGMAGRDT
**:*:***:***** **.**:***** . *: * ****..:* *:*
Mflv_2730|M.gilvum_PYR-GCK GGLSGGELQRLAVAAALAREPSLLVADEVTSMVDQDGREDLMSVLSGLTR
Mvan_3806|M.vanbaalenii_PYR-1 GGLSGGELQRLAVAAALAREPSLLIADEVTSMVDQDGREALMSVLAGLTT
TH_0730|M.thermoresistible__bu GALSGGELQRLAVAAALAREPSLLIADEITSMVDQQGREALMSVLSGLTH
MSMEG_4470|M.smegmatis_MC2_155 GGLSGGELQRLAVAAALAREPSMLIADEITSMVDQQGRETLMNVVSGLTK
Mb2353c|M.bovis_AF2122/97 GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGRDALLAVLSGLTQ
Rv2326c|M.tuberculosis_H37Rv GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGRDALLAVLSGLTQ
MMAR_3626|M.marinum_M GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGREALLRVLSGLTK
MUL_1239|M.ulcerans_Agy99 GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGREALLRVLSGLTK
MLBr_00848|M.leprae_Br4923 GSLSGGELQRLALAAALAREPSLLIADEVTSMVDRQGRDALLGVLSGLTK
MAV_2082|M.avium_104 GSLSGGELQRLALAAALARDPKLLIADEVTTMVDQQGRDALLGILSGLAK
MAB_3144|M.abscessus_ATCC_1997 AGLSGGELQRLAVASALAREPALLIADEVTTMVDHDGRQTLINVLDGLTT
..**********:*:****:* :*:***:*:***::**: *: :: **:
Mflv_2730|M.gilvum_PYR-GCK RHRTALVHITHYNDEADAADRTVTLAGN-GADSDNMEMVETADAPSASEE
Mvan_3806|M.vanbaalenii_PYR-1 RHRTALVHITHYNDEAEAADRTVTLVGN-DAASDNSQMVETAEVPSGTPD
TH_0730|M.thermoresistible__bu RHRMALVHITHYESEAEAADRTIALTGVDDRGTAGGQMVEHVAAPAPSPN
MSMEG_4470|M.smegmatis_MC2_155 RHRMSLVHITHYNSEAEIADRTVALDGD-GGAADSTDMVETSSVHTGS-M
Mb2353c|M.bovis_AF2122/97 RHRTALVHITHYDNEADSADRTLSLSDS----PDNTDMVHTAAMPAPVIG
Rv2326c|M.tuberculosis_H37Rv RHRTALVHITHYDNEADSADRTLSLSDS----PDNTDMVHTAAMPAPVIG
MMAR_3626|M.marinum_M RHRTALVHITHYNNEADSADRTVDLSES----PDNAEMVETAPVPAPAVA
MUL_1239|M.ulcerans_Agy99 RHRTALVHITHYNNEADSADRTVDLSES----PDNAEMVETAPVPAPAVA
MLBr_00848|M.leprae_Br4923 RHPIALVHITHYNNEADTADRTINLSDS----PDNAGMAETVAPPVSTVA
MAV_2082|M.avium_104 RHQTALVHITHYDNEAASADRVIKLSDS----PDNAVAAETNAAAAPAVA
MAB_3144|M.abscessus_ATCC_1997 HHRLGLVHITHYPQEADAADRVVALGGI-----------AVRPPAEPPSP
:* .******* .** ***.: *
Mflv_2730|M.gilvum_PYR-GCK SGPPRGVPVLALRDVGHEYGSGTPWAKTALSGIDFTVHEGDGVLIHGLNG
Mvan_3806|M.vanbaalenii_PYR-1 ETP-RGAPVLELRGVGHEYGSGTPWAKRALSGIDFTVHEGDGVLIHGLNG
TH_0730|M.thermoresistible__bu PGRPDGAPVLQLTGVGHEYGSGTPWAATALRDVDLTIHEGEGVLIHGLNG
MSMEG_4470|M.smegmatis_MC2_155 AGHEPGVPVLELTGVGHEYANGTPWAKTALRDVTFTVNEGDGVLIHGLNG
Mb2353c|M.bovis_AF2122/97 VDQPQHAPALELVGVGHEYASGTPWAKTALRDINFVVEQGDGVLIHGGNG
Rv2326c|M.tuberculosis_H37Rv VDQPQHAPALELVGVGHEYASGTPWAKTALRDINFVVEQGDGVLIHGGNG
MMAR_3626|M.marinum_M VSVGVQRPALELIGVGHEYNIGTPWAKTALRDLNFVVEQGDGVLIHGGNG
MUL_1239|M.ulcerans_Agy99 VSVGVQRPALELIGVGHEYNIGTPWAKTALRDLNFVVEQGDGVLIHGGNG
MLBr_00848|M.leprae_Br4923 VDHRPHAPVLELVGVGHEYGSGTPWAKAALHDISFVVRQGDGVLVYGSNG
MAV_2082|M.avium_104 VPHGSGVPVLELIDVSHEYASGTPWSKVALRDVSFVVEQGDGLLIHGGNG
MAB_3144|M.abscessus_ATCC_1997 AESPSKETILDVRSVSFDYAVGTPWSQPVLRDVSFRVDSGDGILLCGGNG
. * : .*..:* ****: .* .: : : .*:*:*: * **
Mflv_2730|M.gilvum_PYR-GCK SGKSTLAWIMAGLAVPTVGSCLLDGKPVSDQVGAVAISFQAARLQLMRSH
Mvan_3806|M.vanbaalenii_PYR-1 SGKSTLAWIMAGLTVPTVGACLLDGEPVSEQVGAVAISFQAARLQLMRSH
TH_0730|M.thermoresistible__bu SGKSTLAWIMAGLTVPTWGSCLLDGAPVSEQVGAVAISFQAARLQLMRNR
MSMEG_4470|M.smegmatis_MC2_155 SGKSTLAWIMAGLTVPTYGACLLDGAPVSEQVGAVALSFQAARLQLMRSS
Mb2353c|M.bovis_AF2122/97 SGKSTLAWIMAGLTIPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
Rv2326c|M.tuberculosis_H37Rv SGKSTLAWIMAGLTIPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MMAR_3626|M.marinum_M SGKSTLAWVMAGLTVPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MUL_1239|M.ulcerans_Agy99 SGKSTLAWVMAGLTVPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MLBr_00848|M.leprae_Br4923 SGKSTLAWIMAGLMVPTTGACLIDGRPTHEHVGAVALSFQAARLQLMRSR
MAV_2082|M.avium_104 SGKSTLAWIMAGLTTPTSGSCLIDGRPTHERVGEVALSFQSARLQLMRDH
MAB_3144|M.abscessus_ATCC_1997 SGKSTLAWIMAGLLDPSAGQCLLDGRPTAEQVGAVALCFQAARLQLLRGH
********:**** *: * **:** *. ::** **:.**:*****:*.
Mflv_2730|M.gilvum_PYR-GCK VAHEIASAAGFSVRDRDRVITALGTVGLDPALADRRIDQLSGGQMRRVVL
Mvan_3806|M.vanbaalenii_PYR-1 VAHEIASAAGFSVRDRDRVVAALASVGLDPGLATRRIDQLSGGQMRRVVL
TH_0730|M.thermoresistible__bu VDHEIASAAGFPVRDRARVLRALATVGLEPALADRPIDQLSGGQMRRVVL
MSMEG_4470|M.smegmatis_MC2_155 VAREIASAAGFSAKDRDRVNAALATVGLDPTLGERRIDQLSGGQMRRVVL
Mb2353c|M.bovis_AF2122/97 VDLEVASAAGFSASEQDRVAAALTVVGLDPALGARRIDQLSGGQMRRVVL
Rv2326c|M.tuberculosis_H37Rv VDLEVASAAGFSASEQDRVAAALTVVGLDPALGARRIDQLSGGQMRRVVL
MMAR_3626|M.marinum_M VDLEVASAAGFSARDEDRVAAALSVVGLDPTLAKRSIDQLSGGQMRRVVL
MUL_1239|M.ulcerans_Agy99 VDLEVASAAGFSARDEDRVAAALSVVGLDPTLAKRSIDQLSGGQMRRVVL
MLBr_00848|M.leprae_Br4923 VDLEVASAAGFSPRDEDRVAAALGVVGLDPALAKRRIDQLSGGQMRRVVL
MAV_2082|M.avium_104 VDTEVASAAGFSPTDQDRVAEALMSVGLDPAMGKRRIDQLSGGQMRRVVL
MAB_3144|M.abscessus_ATCC_1997 AGAAVAALAGYSAADTDSIDRALASVSLDPAIGGVLIDRLSGGQLRRVAL
. :*: **:. : : ** *.*:* :. **:*****:***.*
Mflv_2730|M.gilvum_PYR-GCK SGLLARSPRALILDEPLAGLDAASQRGLLHLLVDLRRRKRLTVVVISHDF
Mvan_3806|M.vanbaalenii_PYR-1 AGLLARSPRVLILDEPLAGLDAASQRGLVHLLEDLRRRTRLTVVVISHDF
TH_0730|M.thermoresistible__bu AGLLARSPRALILDEPLAGLDAGSQRGLLRILEELR-AAGLTLVVISHDF
MSMEG_4470|M.smegmatis_MC2_155 AGLLARSPRALILDEPLAGLDAASARGLLRLLEDLRRNNGMTVVVISHDF
Mb2353c|M.bovis_AF2122/97 AGLLARAPRALILDEPLAGLDAASQRGLLRLLEDLRRARGLTVVVVSHDF
Rv2326c|M.tuberculosis_H37Rv AGLLARAPRALILDEPLAGLDAASQRGLLRLLEDLRRARGLTVVVVSHDF
MMAR_3626|M.marinum_M AGLLACSPRALILDEPLAGLDATSQRGLLRLLEDLRREQGLTVVVISHDF
MUL_1239|M.ulcerans_Agy99 AGLLACSPRALILDEPLAGLDATSQRGLLRLLEDLRREQGLTVVVISHDF
MLBr_00848|M.leprae_Br4923 AGLLACSPRALILDEPLAGLDAVSQRGLLRLLEDLRCERGLTVVVTSHDF
MAV_2082|M.avium_104 AGLLARSPRALVLDEPLAGLDIGSQRGLLRLLENLRRERGLTVVVISHDT
MAB_3144|M.abscessus_ATCC_1997 AGLLARSPRLLILDEPLAGLDVDAQADLIDLLIRIR-NGGQAVIVISHDT
:**** :** *:********* : .*: :* :* :::* ***
Mflv_2730|M.gilvum_PYR-GCK AGLENLCPRTLHLADGVLATEPTTAGGRS----------
Mvan_3806|M.vanbaalenii_PYR-1 GGLERLCPRTLHLENGVLATAPTAAGGRS----------
TH_0730|M.thermoresistible__bu SGLDGLCTRTVHLENGRVVPARTTAGGMS----------
MSMEG_4470|M.smegmatis_MC2_155 TGLEGLCPRTLHLRDGQLATAPTTAGDAR----------
Mb2353c|M.bovis_AF2122/97 AGMEELCPRTLHLRDGVLESAAASEAGGMS---------
Rv2326c|M.tuberculosis_H37Rv AGMEELCPRTLHLRDGVLESAAASEAGGMS---------
MMAR_3626|M.marinum_M AGMDELCPRTLHLNNGVMEPVSTTAAGGLGGRTGAGGKP
MUL_1239|M.ulcerans_Agy99 AGMDELCPRTLHLNNGVMEPVSTTAAGGLGGRTGAGGKP
MLBr_00848|M.leprae_Br4923 VGLEDVCPRTVHLRNGALESVSTTAGGTS----------
MAV_2082|M.avium_104 VGLEELCPRSLYLRDGALQTASTAAGGMP----------
MAB_3144|M.abscessus_ATCC_1997 DSLSPLCPRTIRLEQGELVTAG-----------------
.:. :*.*:: * :* : .