For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CCAVCGPEPGRIGEVTPLGPCPAQHRGGPLRPSELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTG LLAYRYRLRVLAAATVAAGMIAFLIAGLGGFMGVVHSAYIGGLTGIVKRRGRGTPTVVVSSLIGGFVFGA AMVGMLAAMVRLRHLIFKVMTANVDGIAATLARMHMQGAAADVKRYFAEGLQYWPWVLLGYFNIGIMIVS LIGWWALSRLLERMRGIPDVHKLDPPPGDDVDALIGPVPVRLDKVRFRYPRAGQDALREVSLDVRAGEHL AIIGANGSGKTTLMLILAGRAPTSGTVDRPGTVGLGKLGGTAVVLQHPESQVLGTRVADDVVWGLPLGTT ADVGRLLSEVGLEALAERDTGSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGRDALLAVLSGLTQ RHRTALVHITHYDNEADSADRTLSLSDSPDNTDMVHTAAMPAPVIGVDQPQHAPALELVGVGHEYASGTP WAKTALRDINFVVEQGDGVLIHGGNGSGKSTLAWIMAGLTIPTTGACLLDGRPTHEQVGAVALSFQAARL QLMRSRVDLEVASAAGFSASEQDRVAAALTVVGLDPALGARRIDQLSGGQMRRVVLAGLLARAPRALILD EPLAGLDAASQRGLLRLLEDLRRARGLTVVVVSHDFAGMEELCPRTLHLRDGVLESAAASEAGGMS*
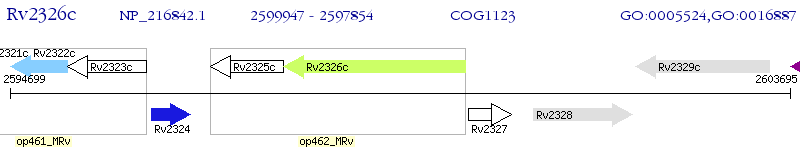
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2326c | - | - | 100% (697) | POSSIBLE TRANSMEMBRANE ATP-BINDING PROTEIN ABC TRANSORTER |
| M. tuberculosis H37Rv | Rv3663c | dppD | 2e-33 | 29.44% (496) | peptide ABC transporter ATP-binding protein |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 6e-19 | 34.33% (201) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2353c | - | 0.0 | 100.00% (697) | putative transmembrane ATP-binding protein ABC transorter |
| M. gilvum PYR-GCK | Mflv_2730 | - | 0.0 | 65.43% (703) | ABC transporter related |
| M. leprae Br4923 | MLBr_00848 | - | 0.0 | 77.39% (690) | ABC transporter |
| M. abscessus ATCC 19977 | MAB_1713 | - | 0.0 | 58.88% (659) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_3626 | - | 0.0 | 83.55% (681) | transmembrane ATP-binding protein ABC transorter |
| M. avium 104 | MAV_2082 | - | 0.0 | 72.52% (655) | ABC transporter |
| M. smegmatis MC2 155 | MSMEG_4470 | - | 0.0 | 70.09% (672) | ABC transporter |
| M. thermoresistible (build 8) | TH_0730 | - | 0.0 | 66.23% (687) | POSSIBLE TRANSMEMBRANE ATP-BINDING PROTEIN ABC TRANSORTER |
| M. ulcerans Agy99 | MUL_1239 | - | 0.0 | 83.55% (681) | transmembrane ATP-binding protein ABC transorter |
| M. vanbaalenii PYR-1 | Mvan_3806 | - | 0.0 | 69.53% (686) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2326c|M.tuberculosis_H37Rv ----------------------------MCCAVCGPEPGRIGEVTPLGPC
Mb2353c|M.bovis_AF2122/97 ----------------------------MCCAVCGPEPGRIGEVTPLGPC
MMAR_3626|M.marinum_M -------------------------------------------MTASAPL
MUL_1239|M.ulcerans_Agy99 -------------------------------------------MTASAPL
MLBr_00848|M.leprae_Br4923 MDLCIHRRMALLCQATRRCHRWKTQLGRCAAPDLAPGTGTIGKVTAHDPR
MAV_2082|M.avium_104 --------------------------------------------------
Mflv_2730|M.gilvum_PYR-GCK ----------------MSAAVRPTSPPEAGRAGQFSGDWQNREMTSGRSE
Mvan_3806|M.vanbaalenii_PYR-1 -------------------------------------------MTSAGPE
MSMEG_4470|M.smegmatis_MC2_155 -------------------------------------------MTANRPA
TH_0730|M.thermoresistible__bu -------------------------------------------MTMTGQA
MAB_1713|M.abscessus_ATCC_1997 --------------------------------------------------
Rv2326c|M.tuberculosis_H37Rv PAQHRGGPLRPSELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
Mb2353c|M.bovis_AF2122/97 PAQHRGGPLRPSELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MMAR_3626|M.marinum_M APPHRGGPLRPGELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MUL_1239|M.ulcerans_Agy99 APPHRGGPLRPGELAQASVMAALCAVTAIISVVVPFAAGLALLGTVPTGL
MLBr_00848|M.leprae_Br4923 HLHHRSGSLQPGELAQASVLAALCAVTAIVSVVVPFAAGLALLGTVPMGL
MAV_2082|M.avium_104 -------------------MGALCAAIEILAAVIPFAQGLGVLGTVPMGL
Mflv_2730|M.gilvum_PYR-GCK SDTRRTGRLQPVELAQAAVMAALCAAIAIIAVVVPFAAGLSLVGTVPMGL
Mvan_3806|M.vanbaalenii_PYR-1 SAPRRTGPLQPVELAQAAVMAALCAAIAIIAVVVPFAGSLSLLGTVPMGL
MSMEG_4470|M.smegmatis_MC2_155 RAPRRG--LTPGELAQASVMAALCAATAIIAVVVPFAAGLSVLGTVPMGL
TH_0730|M.thermoresistible__bu RRPHRAGSLHPVELAHASVMAALTAATAIIAVVVPFAAGVSLLGTVPMGM
MAB_1713|M.abscessus_ATCC_1997 -MNFARVPLKPDELAQAAVMAALTAAMVIVGIALPILGAVQLLGAVPMGL
:.** *. *:. .:*: .: ::*:** *:
Rv2326c|M.tuberculosis_H37Rv LAYRYRLRVLAAATVAAGMIAFLIAGLGGFMGVVHSAYIGGLTGIVKRRG
Mb2353c|M.bovis_AF2122/97 LAYRYRLRVLAAATVAAGMIAFLIAGLGGFMGVVHSAYIGGLTGIVKRRG
MMAR_3626|M.marinum_M LAYRYRLRVLVAATVAAGVIAFLIAGLGGFMGVVHSAYIGGLTGIVKRKG
MUL_1239|M.ulcerans_Agy99 LAYRYRLRVLVAATVAAGVIAFLIAGLGGFMGVVHSAYIGGLTGIVKRKG
MLBr_00848|M.leprae_Br4923 LAYRYRFRVLMTAMVAAGVIAFLITGLGGFIAVVNSAYIGGLTGVVKRKG
MAV_2082|M.avium_104 LAYRYRPRALMAATVAGGVIAFLIAGLGSLFMLVDCAWVGGLCGIVKRKG
Mflv_2730|M.gilvum_PYR-GCK LAYRHRMRVLLAATVAGGLVAFLIAGMGGLMTVINCAYIGGLTGIVKRRG
Mvan_3806|M.vanbaalenii_PYR-1 LAYRYRLRVLLAATVAGGLVAFLIAGMGGFMTVVNCAYIGGLTGIVKRRG
MSMEG_4470|M.smegmatis_MC2_155 LAYRYRLRVLLAATVAAGVIAFLIAGLGGMMTVFNCAYIGGLTGIVKRRG
TH_0730|M.thermoresistible__bu LAYRFRFRVLIAATVAGATIAFLIAGMGGFMTVVNCAYIGGLTGTVKRRG
MAB_1713|M.abscessus_ATCC_1997 LGYRYRIRVLLAAAVAGGVIAFLLAGLGGFIAMCQCVYIGGLTGVVRRRG
*.**.* *.* :* **.. :***::*:*.:: : ...::*** * *:*:*
Rv2326c|M.tuberculosis_H37Rv RGTPTVVVSSLIGGFVFGAAMVGMLAAMVRLRHLIFKVMTANVDGIAATL
Mb2353c|M.bovis_AF2122/97 RGTPTVVVSSLIGGFVFGAAMVGMLAAMVRLRHLIFKVMTANVDGIAATL
MMAR_3626|M.marinum_M RGTPTVIVSSLIAGLAFGSAMVGMLAALTRLRHLIFKVMTANVEGLSSFM
MUL_1239|M.ulcerans_Agy99 RGTPTVIVSSLIAGLAFGSAMVGMLAALTRLRHLIFKVMTANVEGLSSFM
MLBr_00848|M.leprae_Br4923 QGTLTVIALALFAGLAFGAANVVALVVLGRLRHLIFKAMTANVDGIAATL
MAV_2082|M.avium_104 RGTPTVALLSLIAGVLWGAGWVAVLAVLTRLRHLFFDVITANANGVAAFL
Mflv_2730|M.gilvum_PYR-GCK RGTPTVFVCALVAGALFGIVIVAALGVLSRLRNLIFDSVTANIAGTAAVL
Mvan_3806|M.vanbaalenii_PYR-1 RGTPTVVACALVAGALFGVAVVAALALLSRLRHLIFDSMTANITGLAAIL
MSMEG_4470|M.smegmatis_MC2_155 RGTPTVFAAAIVAGALFGAMIVVALLVLSRLRNLMFDTLTANIDGLAAVF
TH_0730|M.thermoresistible__bu RGTGTVLVAALIAGAAFGAAIVGALTVLSRLRHLMFDTITANVHGVARII
MAB_1713|M.abscessus_ATCC_1997 HGMPLLTVIGVICSAIIAALAVGALTLLNRMRTLLFDALRASLEGLFKMI
:* : .:. . . * * : *:* *:*. : *. * :
Rv2326c|M.tuberculosis_H37Rv ARMH-MQGAAADVKRYFAEGLQYWPWVLLGYFNIGIMIVSLIGWWALSRL
Mb2353c|M.bovis_AF2122/97 ARMH-MQGAAADVKRYFAEGLQYWPWVLLGYFNIGIMIVSLIGWWALSRL
MMAR_3626|M.marinum_M ARMG-MEGAAEDLKRYFAEGLHYWPWVLLGYFTVAIMVVSLIGWWALSRL
MUL_1239|M.ulcerans_Agy99 ARMG-MEGAAEDLKRYFAEGLHYWPWVLLGYFTVAIMVVSLIGWWALSRL
MLBr_00848|M.leprae_Br4923 TWMH-LPWVAVQLKRYFADGLQHWPWMLLGYFVITILVVSLIGWWVLSRV
MAV_2082|M.avium_104 NWMH-LQGVGAGLKRYVADGLQHWPLLIFPYMILLVVVVSFISWSALSRL
Mflv_2730|M.gilvum_PYR-GCK ERVPGLAPVGERLTRDFATMLHYWPFLFFASGVVSITFVSLVGWWALSRV
Mvan_3806|M.vanbaalenii_PYR-1 ERVPGLEPVAERLTRDFATALHYWPYLFFAVGVLSITFVSLVGWWALSRV
MSMEG_4470|M.smegmatis_MC2_155 ARVPVLEGAAEPLKTMFATLLHHWPVLMLLMGISSITMVTMVGWWALSRV
TH_0730|M.thermoresistible__bu SGIPNMSGAGERLRRDFETMLSYWPVLILAAGVISITVVTLIGWWALSRV
MAB_1713|M.abscessus_ATCC_1997 GHIPHLEAVGVNATSAIDYAMRNWMWFIGGIVFTFVLLNTMFGWWVLSRV
: : .. . : * .: : . ::..* .***:
Rv2326c|M.tuberculosis_H37Rv LERMRGIPDVHKLDPPPGDDVDALIGPVPVRLDKVRFRYPRAGQDALREV
Mb2353c|M.bovis_AF2122/97 LERMRGIPDVHKLDPPPGDDVDALIGPVPVRLDKVRFRYPRAGQDALREV
MMAR_3626|M.marinum_M LERMRGIPDVHKLDAP-VDGEATSIEPVPVRLDKVRFRYPGAGQDALREV
MUL_1239|M.ulcerans_Agy99 LERMRGIPDVHKLDAP-VDGEATSIEPVPVRLDKVRFRYPGAGQDALREV
MLBr_00848|M.leprae_Br4923 LERIRDIPDVHKLDAPSACNEDAPVGPVPVWLDKVRFRYPHAGQDALREV
MAV_2082|M.avium_104 LDRMRKIPDVHKLDPP--DDGHAAIGPVPVRLENVRFRYPGAEQDALREV
Mflv_2730|M.gilvum_PYR-GCK LKRLAGVPDVHKLEAS---ADTGEIAPTPLTLTDVRFRYPGTDHDALGPV
Mvan_3806|M.vanbaalenii_PYR-1 LGRLLGVPDVHKLEAS---NDVGSIAPVPLQLRDVRFRYPNAGQDALGPV
MSMEG_4470|M.smegmatis_MC2_155 LGRLLGVPDVHKLDSP---PEHGPIGPVPAQLTDVRFRYPGADHDALGPV
TH_0730|M.thermoresistible__bu LARLVVIPDVHKLESF---EDTGPVAPVPLRLRDVRFRYPNADRDALGPL
MAB_1713|M.abscessus_ATCC_1997 IRRLSVIPDVHQLDPP---AVAGAVGPVPARLHEVRFRYPRGESDALAPL
: *: :****:*:. : *.* * .****** *** :
Rv2326c|M.tuberculosis_H37Rv SLDVRAGEHLAIIGANGSGKTTLMLILAGRAPTSGTVDRPGTVGLGKLGG
Mb2353c|M.bovis_AF2122/97 SLDVRAGEHLAIIGANGSGKTTLMLILAGRAPTSGTVDRPGTVGLGKLGG
MMAR_3626|M.marinum_M SLDVRVGEHIAITGANGSGKTTLMLILAGREPTSGTVERPGAVGLGKLGG
MUL_1239|M.ulcerans_Agy99 SLDVRVGEHIAITGANGSGKTTLMLILAGREPTSGTVERPGAVGLGKLGG
MLBr_00848|M.leprae_Br4923 SLDLRVGEHVAVTGANGSGKTTLMLILAGREPTSGTVDRPGAVGLGKLGG
MAV_2082|M.avium_104 SLDIQAGEHVAVTGANGSGKTTLMLILAGRQPTSGTVHRPGAVGLGDVDG
Mflv_2730|M.gilvum_PYR-GCK SLAVQPGEHVAVTGANGSGKTTLMLVLSGREPTGGTVERPGAVGLGRIGG
Mvan_3806|M.vanbaalenii_PYR-1 SLAVQPGEHLAVTGANGSGKTTLMLVLSGREATAGTVERPGAVGLGKIGG
MSMEG_4470|M.smegmatis_MC2_155 TMDLRVGEHVAVTGPNGSGKTTLMLVLAGREPTSGSVERAGAVGLGRMGG
TH_0730|M.thermoresistible__bu TMTVQPGEHVAITGANGSGKTTLMLLLAGREPTAGTVERPGAVGLGRMGG
MAB_1713|M.abscessus_ATCC_1997 SMNIDVGEHVAITGPNGSGKSTLMRILAGREPTSGQIERPGSVGLGAIGG
:: : ***:*: *.*****:*** :*:** .*.* :.*.*:**** :.*
Rv2326c|M.tuberculosis_H37Rv TAVVLQHPESQVLGTRVADDVVWGLPLGTTADVGRLLSEVGLEALAERDT
Mb2353c|M.bovis_AF2122/97 TAVVLQHPESQVLGTRVADDVVWGLPLGTTADVGRLLSEVGLEALAERDT
MMAR_3626|M.marinum_M TAVVLQHPESQVLGTRVADDVVWGLPPGTKIDVDQQLAEVGLEGLAERET
MUL_1239|M.ulcerans_Agy99 TAVVLQHPESQVLGTRVADDVVWGLPPGTKIDVDQQLAEVGLEGLAERET
MLBr_00848|M.leprae_Br4923 TAVVLQHPESQVLGTRVADDVVWGLPPGTDVDVNRLLREVGLDAFAERDT
MAV_2082|M.avium_104 TAIVLQHPKSQVLGTRVADDVVWGLPPGTDIDVHRLLREVGLDGLAERDT
Mflv_2730|M.gilvum_PYR-GCK TAVVMQHPESQVLGTRVADDLVWGLPPGTAVDVDRLLGEVGLDGLAERDT
Mvan_3806|M.vanbaalenii_PYR-1 TAVVMQHPESQVLGTRVADDVVWGLPPGASIDVDRLLAEVGLDGLAERDT
MSMEG_4470|M.smegmatis_MC2_155 TAVVMQHPESQVLGTRVADDVVWGLPPGTATDVDRLLSEVGLDGMAERDT
TH_0730|M.thermoresistible__bu TAVVMQHPESQVLGTRVADDVVWGLPPGTATDMPRLLEEVGLDGLAERDT
MAB_1713|M.abscessus_ATCC_1997 TAIVMQHPESQVLGTKVADDVVWGLPADHKVDIGGLLREVGLEGMAEHST
**:*:***:******:****:***** . *: * ****:.:**:.*
Rv2326c|M.tuberculosis_H37Rv GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGRDALLAVLSGLTQ
Mb2353c|M.bovis_AF2122/97 GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGRDALLAVLSGLTQ
MMAR_3626|M.marinum_M GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGREALLRVLSGLTK
MUL_1239|M.ulcerans_Agy99 GSLSGGELQRLALAAALAREPAMLIADEVTTMVDQQGREALLRVLSGLTK
MLBr_00848|M.leprae_Br4923 GSLSGGELQRLALAAALAREPSLLIADEVTSMVDRQGRDALLGVLSGLTK
MAV_2082|M.avium_104 GSLSGGELQRLALAAALARDPKLLIADEVTTMVDQQGRDALLGILSGLAK
Mflv_2730|M.gilvum_PYR-GCK GGLSGGELQRLAVAAALAREPSLLVADEVTSMVDQDGREDLMSVLSGLTR
Mvan_3806|M.vanbaalenii_PYR-1 GGLSGGELQRLAVAAALAREPSLLIADEVTSMVDQDGREALMSVLAGLTT
MSMEG_4470|M.smegmatis_MC2_155 GGLSGGELQRLAVAAALAREPSMLIADEITSMVDQQGRETLMNVVSGLTK
TH_0730|M.thermoresistible__bu GALSGGELQRLAVAAALAREPSLLIADEITSMVDQQGREALMSVLSGLTH
MAB_1713|M.abscessus_ATCC_1997 SSLSGGELQRLAVASALARDPALLVADEITSMVDPQGRQDLLGVLSRLTQ
..**********:*:****:* :*:***:*:*** :**: *: ::: *:
Rv2326c|M.tuberculosis_H37Rv RHRTALVHITHYDNEADSADRTLSLSD----SPDNTDMVHTAAMPAPVIG
Mb2353c|M.bovis_AF2122/97 RHRTALVHITHYDNEADSADRTLSLSD----SPDNTDMVHTAAMPAPVIG
MMAR_3626|M.marinum_M RHRTALVHITHYNNEADSADRTVDLSE----SPDNAEMVETAPVPAPAVA
MUL_1239|M.ulcerans_Agy99 RHRTALVHITHYNNEADSADRTVDLSE----SPDNAEMVETAPVPAPAVA
MLBr_00848|M.leprae_Br4923 RHPIALVHITHYNNEADTADRTINLSD----SPDNAGMAETVAPPVSTVA
MAV_2082|M.avium_104 RHQTALVHITHYDNEAASADRVIKLSD----SPDNAVAAETNAAAAPAVA
Mflv_2730|M.gilvum_PYR-GCK RHRTALVHITHYNDEADAADRTVTLAG-NGADSDNMEMVETADAPSASEE
Mvan_3806|M.vanbaalenii_PYR-1 RHRTALVHITHYNDEAEAADRTVTLVG-NDAASDNSQMVETAEVPSGTPD
MSMEG_4470|M.smegmatis_MC2_155 RHRMSLVHITHYNSEAEIADRTVALDG-DGGAADSTDMVETSSVHTGS-M
TH_0730|M.thermoresistible__bu RHRMALVHITHYESEAEAADRTIALTGVDDRGTAGGQMVEHVAAPAPSPN
MAB_1713|M.abscessus_ATCC_1997 QHRMSLVHITHFQSEAAAADREVALRAVN--GKAARTAIQTVPVPATSAA
:* :******::.** *** : * .
Rv2326c|M.tuberculosis_H37Rv VDQPQHAPALELVGVGHEYASGTPWAKTALRDINFVVEQGDGVLIHGGNG
Mb2353c|M.bovis_AF2122/97 VDQPQHAPALELVGVGHEYASGTPWAKTALRDINFVVEQGDGVLIHGGNG
MMAR_3626|M.marinum_M VSVGVQRPALELIGVGHEYNIGTPWAKTALRDLNFVVEQGDGVLIHGGNG
MUL_1239|M.ulcerans_Agy99 VSVGVQRPALELIGVGHEYNIGTPWAKTALRDLNFVVEQGDGVLIHGGNG
MLBr_00848|M.leprae_Br4923 VDHRPHAPVLELVGVGHEYGSGTPWAKAALHDISFVVRQGDGVLVYGSNG
MAV_2082|M.avium_104 VPHGSGVPVLELIDVSHEYASGTPWSKVALRDVSFVVEQGDGLLIHGGNG
Mflv_2730|M.gilvum_PYR-GCK SGPPRGVPVLALRDVGHEYGSGTPWAKTALSGIDFTVHEGDGVLIHGLNG
Mvan_3806|M.vanbaalenii_PYR-1 ETP-RGAPVLELRGVGHEYGSGTPWAKRALSGIDFTVHEGDGVLIHGLNG
MSMEG_4470|M.smegmatis_MC2_155 AGHEPGVPVLELTGVGHEYANGTPWAKTALRDVTFTVNEGDGVLIHGLNG
TH_0730|M.thermoresistible__bu PGRPDGAPVLQLTGVGHEYGSGTPWAATALRDVDLTIHEGEGVLIHGLNG
MAB_1713|M.abscessus_ATCC_1997 RPDNESPVVLEFDSVGHEYASGTPWAKTALRDVSLAVHEGDGVLIHGGNG
.* : .*.*** ****: ** .: :.:.:*:*:*::* **
Rv2326c|M.tuberculosis_H37Rv SGKSTLAWIMAGLTIPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
Mb2353c|M.bovis_AF2122/97 SGKSTLAWIMAGLTIPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MMAR_3626|M.marinum_M SGKSTLAWVMAGLTVPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MUL_1239|M.ulcerans_Agy99 SGKSTLAWVMAGLTVPTTGACLLDGRPTHEQVGAVALSFQAARLQLMRSR
MLBr_00848|M.leprae_Br4923 SGKSTLAWIMAGLMVPTTGACLIDGRPTHEHVGAVALSFQAARLQLMRSR
MAV_2082|M.avium_104 SGKSTLAWIMAGLTTPTSGSCLIDGRPTHERVGEVALSFQSARLQLMRDH
Mflv_2730|M.gilvum_PYR-GCK SGKSTLAWIMAGLAVPTVGSCLLDGKPVSDQVGAVAISFQAARLQLMRSH
Mvan_3806|M.vanbaalenii_PYR-1 SGKSTLAWIMAGLTVPTVGACLLDGEPVSEQVGAVAISFQAARLQLMRSH
MSMEG_4470|M.smegmatis_MC2_155 SGKSTLAWIMAGLTVPTYGACLLDGAPVSEQVGAVALSFQAARLQLMRSS
TH_0730|M.thermoresistible__bu SGKSTLAWIMAGLTVPTWGSCLLDGAPVSEQVGAVAISFQAARLQLMRNR
MAB_1713|M.abscessus_ATCC_1997 SGKSTFAWIAAGLTKPTTGQCLLDGRPVSDQVGSVAISFQAARLQLLRSR
*****:**: *** ** * **:** *. ::** **:***:*****:*.
Rv2326c|M.tuberculosis_H37Rv VDLEVASAAGFSASEQDRVAAALTVVGLDPALGARRIDQLSGGQMRRVVL
Mb2353c|M.bovis_AF2122/97 VDLEVASAAGFSASEQDRVAAALTVVGLDPALGARRIDQLSGGQMRRVVL
MMAR_3626|M.marinum_M VDLEVASAAGFSARDEDRVAAALSVVGLDPTLAKRSIDQLSGGQMRRVVL
MUL_1239|M.ulcerans_Agy99 VDLEVASAAGFSARDEDRVAAALSVVGLDPTLAKRSIDQLSGGQMRRVVL
MLBr_00848|M.leprae_Br4923 VDLEVASAAGFSPRDEDRVAAALGVVGLDPALAKRRIDQLSGGQMRRVVL
MAV_2082|M.avium_104 VDTEVASAAGFSPTDQDRVAEALMSVGLDPAMGKRRIDQLSGGQMRRVVL
Mflv_2730|M.gilvum_PYR-GCK VAHEIASAAGFSVRDRDRVITALGTVGLDPALADRRIDQLSGGQMRRVVL
Mvan_3806|M.vanbaalenii_PYR-1 VAHEIASAAGFSVRDRDRVVAALASVGLDPGLATRRIDQLSGGQMRRVVL
MSMEG_4470|M.smegmatis_MC2_155 VAREIASAAGFSAKDRDRVNAALATVGLDPTLGERRIDQLSGGQMRRVVL
TH_0730|M.thermoresistible__bu VDHEIASAAGFPVRDRARVLRALATVGLEPALADRPIDQLSGGQMRRVVL
MAB_1713|M.abscessus_ATCC_1997 VDYEIASAAGFSARDTDRVVRALASVGLEPDLATRPIDQLSGGQMRRVVL
* *:******. : ** ** ***:* :. * **************
Rv2326c|M.tuberculosis_H37Rv AGLLARAPRALILDEPLAGLDAASQRGLLRLLEDLRRARGLTVVVVSHDF
Mb2353c|M.bovis_AF2122/97 AGLLARAPRALILDEPLAGLDAASQRGLLRLLEDLRRARGLTVVVVSHDF
MMAR_3626|M.marinum_M AGLLACSPRALILDEPLAGLDATSQRGLLRLLEDLRREQGLTVVVISHDF
MUL_1239|M.ulcerans_Agy99 AGLLACSPRALILDEPLAGLDATSQRGLLRLLEDLRREQGLTVVVISHDF
MLBr_00848|M.leprae_Br4923 AGLLACSPRALILDEPLAGLDAVSQRGLLRLLEDLRCERGLTVVVTSHDF
MAV_2082|M.avium_104 AGLLARSPRALVLDEPLAGLDIGSQRGLLRLLENLRRERGLTVVVISHDT
Mflv_2730|M.gilvum_PYR-GCK SGLLARSPRALILDEPLAGLDAASQRGLLHLLVDLRRRKRLTVVVISHDF
Mvan_3806|M.vanbaalenii_PYR-1 AGLLARSPRVLILDEPLAGLDAASQRGLVHLLEDLRRRTRLTVVVISHDF
MSMEG_4470|M.smegmatis_MC2_155 AGLLARSPRALILDEPLAGLDAASARGLLRLLEDLRRNNGMTVVVISHDF
TH_0730|M.thermoresistible__bu AGLLARSPRALILDEPLAGLDAGSQRGLLRILEELR-AAGLTLVVISHDF
MAB_1713|M.abscessus_ATCC_1997 AGLLARSPRALILDEPLAGLDAEAQAGLLRLLVEVRERERLTIVVISHDF
:**** :**.*:********* : **:::* ::* :*:** ***
Rv2326c|M.tuberculosis_H37Rv AGMEELCPRTLHLRDGVLESAAASEAGGMS---------
Mb2353c|M.bovis_AF2122/97 AGMEELCPRTLHLRDGVLESAAASEAGGMS---------
MMAR_3626|M.marinum_M AGMDELCPRTLHLNNGVMEPVSTTAAGGLGGRTGAGGKP
MUL_1239|M.ulcerans_Agy99 AGMDELCPRTLHLNNGVMEPVSTTAAGGLGGRTGAGGKP
MLBr_00848|M.leprae_Br4923 VGLEDVCPRTVHLRNGALESVSTTAGGTS----------
MAV_2082|M.avium_104 VGLEELCPRSLYLRDGALQTASTAAGGMP----------
Mflv_2730|M.gilvum_PYR-GCK AGLENLCPRTLHLADGVLATEPTTAGGRS----------
Mvan_3806|M.vanbaalenii_PYR-1 GGLERLCPRTLHLENGVLATAPTAAGGRS----------
MSMEG_4470|M.smegmatis_MC2_155 TGLEGLCPRTLHLRDGQLATAPTTAGDAR----------
TH_0730|M.thermoresistible__bu SGLDGLCTRTVHLENGRVVPARTTAGGMS----------
MAB_1713|M.abscessus_ATCC_1997 DGMQELCPRTVHLQSGVLLPEAVGSGRRG----------
*:: :*.*:::* .* : . . .