For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMASPSNTTDIRYLERLWVPWWWALPGFGAATLLGLEVNQGLRQLPSWVPYPILFAIVAGVLLWFSRIRV EVAAGPDGTPELRAGSAHLPVSVIAKSAAIPATAKSAALGRQLDPAAFVVHRAWIGPMVLVVLNDADDPT PYWLVSSRHPDRLLAALRS
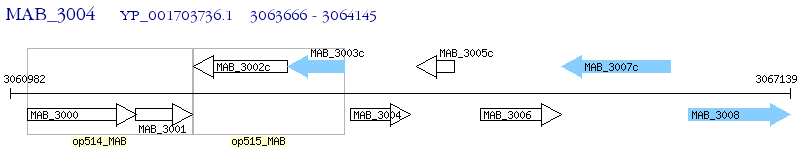
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3004 | - | - | 100% (159) | hypothetical protein MAB_3004 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2717 | - | 3e-55 | 65.56% (151) | alanine rich transmembrane protein |
| M. gilvum PYR-GCK | Mflv_3934 | - | 4e-53 | 60.40% (149) | putative alanine rich transmembrane protein |
| M. tuberculosis H37Rv | Rv2698 | - | 4e-55 | 65.56% (151) | alanine rich transmembrane protein |
| M. leprae Br4923 | MLBr_01027 | - | 4e-52 | 59.01% (161) | hypothetical protein MLBr_01027 |
| M. marinum M | MMAR_2016 | - | 2e-57 | 65.77% (149) | hypothetical protein MMAR_2016 |
| M. avium 104 | MAV_3590 | - | 5e-50 | 61.81% (144) | hypothetical protein MAV_3590 |
| M. smegmatis MC2 155 | MSMEG_2764 | - | 3e-56 | 63.64% (154) | alanine rich transmembrane protein |
| M. thermoresistible (build 8) | TH_1191 | - | 4e-49 | 65.00% (140) | PROBABLE CONSERVED ALANINE RICH TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3336 | - | 2e-57 | 65.77% (149) | hypothetical protein MUL_3336 |
| M. vanbaalenii PYR-1 | Mvan_2466 | - | 8e-53 | 59.73% (149) | putative alanine rich transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2016|M.marinum_M MSETRVAPHSVRYRERLWVPWWWWPLGFALAGLIAFEVNLGIAALADWLP
MUL_3336|M.ulcerans_Agy99 MSETRVAPHSVRYRERLWVPWWWWPLGFALAGLIAFEVNLGIAALADWLP
Mb2717|M.bovis_AF2122/97 MSGTRLAPHSVRYRERLWVPWWWWPLAFALAALIAFEVNLGVAALPDWVP
Rv2698|M.tuberculosis_H37Rv MSGTRLAPHSVRYRERLWVPWWWWPLAFALAALIAFEVNLGVAALPDWVP
MLBr_01027|M.leprae_Br4923 MSGTPVAPHNVRYRERLWVPWWWWPLAFALASLIAFEVNLSGATLPSWLP
MAV_3590|M.avium_104 ----------------MWVPWWWWPPAFALAGIIAFEFNMGVTRQSSWWP
Mflv_3934|M.gilvum_PYR-GCK MSDTRATARTVRYRERLWVPLWWWLPGLGLAALIALEVDQGVRAVPDWLP
Mvan_2466|M.vanbaalenii_PYR-1 MSDTRATAQTVRYRERLTVPLWWWLPGLGVAALIALEINQGIRALPGWLP
TH_1191|M.thermoresistible__bu ------------------VSWWWWPPGLGLAALMAATVTQAAPELPVWLP
MSMEG_2764|M.smegmatis_MC2_155 MSDTRATTQTTRYRERLWVPWWWIPIAAGFAALIAFEVVMGVSSIPAWVP
MAB_3004|M.abscessus_ATCC_1997 MMASPSNTTDIRYLERLWVPWWWALPGFGAATLLGLEVNQGLRQLPSWVP
*. ** . . * ::. . . . * *
MMAR_2016|M.marinum_M YAILFTVAAAALLWLGRVEIRVIAG-ADGE-VELWAGEAHLPVTVIARSA
MUL_3336|M.ulcerans_Agy99 YAILFTVAAAALLWLGRVEIRVIAG-ADGE-VELWAGEAHLPVTVIARSA
Mb2717|M.bovis_AF2122/97 FATLFTVAAGTLLWLGRVEIRVTAGSADGAGVKLWAGPAHLPVAVIARSA
Rv2698|M.tuberculosis_H37Rv FATLFTVAAGTLLWLGRVEIRVTAGSADGAGVKLWAGPAHLPVAVIARSA
MLBr_01027|M.leprae_Br4923 ----FAVAAGTLLWLGRVEIQVIADAPLGGSVELWAGNAHLPITAIAQSA
MAV_3590|M.avium_104 YAALFAVAAAALLWLGRIEVRVTANPAVPGEVELWAGPAHLPVTAIARSA
Mflv_3934|M.gilvum_PYR-GCK YAVLLPVAAVVLIGLGRTEVQVVGD-EGNT--ELWVGNAHLPASVVTRSA
Mvan_2466|M.vanbaalenii_PYR-1 YAVLLPVAAVVLLGLGRTELRVVTAADGQT--ELWVGNAHLPTAVISRTA
TH_1191|M.thermoresistible__bu YAVLTAVAAVVLLWLGRFEVRVVGTGPDDT--ELWVGDAHLPVSVISRMA
MSMEG_2764|M.smegmatis_MC2_155 YAVLLPVAAAVLLWFSKTEVRVVSSPDGDN--ELWVNAAHLPRSVIARSA
MAB_3004|M.abscessus_ATCC_1997 YPILFAIVAGVLLWFSRIRVEVAAGPDGTP--ELRAGSAHLPVSVIAKSA
.:.* .*: :.: .:.* :* .. **** :.::: *
MMAR_2016|M.marinum_M EVPRTAKSAALGRQLDPAAYVLHRAWVGPMVLLVLDDPDDPTPYWLVSCR
MUL_3336|M.ulcerans_Agy99 EVPRTAKSAALGRQLDPAAYVLHRAWVGPMVLLVLDDPDDPTPYWLVSCR
Mb2717|M.bovis_AF2122/97 EIPATAKSAALGRQLDPAAYVLHRAWVGPMVLVVLDDPNDPTPYWLVSCR
Rv2698|M.tuberculosis_H37Rv EIPATAKSAALGRQLDPAAYVLHRAWVGPMVLVVLDDPNDPTPYWLVSCR
MLBr_01027|M.leprae_Br4923 AISRSAKSAALGRQLDPAAYVLHRAWVGPMILVVLDDPDDPTPYWLVSCR
MAV_3590|M.avium_104 PVAPSAKSAALGRQLDPAAYVLHRAWVGPMILVVLDDPDDPTPYWLVSCR
Mflv_3934|M.gilvum_PYR-GCK VVPRSAKSAALGRQLDPAAYVVHKTWVGPLVLLVLDDPDDPTPYWLISAR
Mvan_2466|M.vanbaalenii_PYR-1 EVPRSAKSAALGRQLDPAAYVVHRGWVGPMALLVLDDPDDPTPYWLISTK
TH_1191|M.thermoresistible__bu EVPASAKSAALGRQLDPAAYVVHRPWVGPMVLVVLDDPDDPTPYWLVSTR
MSMEG_2764|M.smegmatis_MC2_155 EVPKSAKSAALGRQLDPAAYVVHRAWVGPMVLVVLDDPDDPTPYWLVSTR
MAB_3004|M.abscessus_ATCC_1997 AIPATAKSAALGRQLDPAAFVVHRAWIGPMVLVVLNDADDPTPYWLVSSR
:. :**************:*:*: *:**: *:**:*.:*******:* :
MMAR_2016|M.marinum_M HPERVLSALRS---
MUL_3336|M.ulcerans_Agy99 HPERVLSALRS---
Mb2717|M.bovis_AF2122/97 HPERVLSALRS---
Rv2698|M.tuberculosis_H37Rv HPERVLSALRS---
MLBr_01027|M.leprae_Br4923 HPERVLSALRS---
MAV_3590|M.avium_104 HPERVLSALRG---
Mflv_3934|M.gilvum_PYR-GCK HPDKVLAALNS---
Mvan_2466|M.vanbaalenii_PYR-1 HPDKVLAALRA---
TH_1191|M.thermoresistible__bu HPDRVLAALGGDSG
MSMEG_2764|M.smegmatis_MC2_155 HPDRLLAALNA---
MAB_3004|M.abscessus_ATCC_1997 HPDRLLAALRS---
**:::*:** .