For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEASSGRKVVALDVDVVRTPANQPLPQRLLVISEVVEHWMETHRPDVVAIERVFAQQNVSTVMGTAQAGG VIALCAAKRGIEVYFHTPSEVKAAVTGNGRADKSQVTAMVTRILNLQAAPKPADAADALALAICHCWRAP MLARMAKAEAVAEEQRKAFAARMKAAAR*
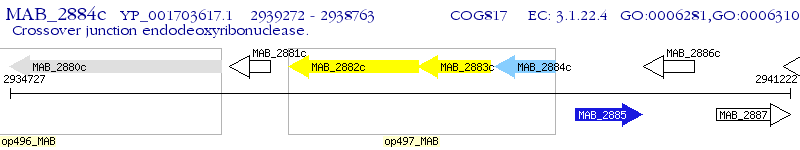
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2884c | - | - | 100% (169) | crossover junction endodeoxyribonuclease RuvC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2625c | ruvC | 1e-66 | 71.93% (171) | Holliday junction resolvase |
| M. gilvum PYR-GCK | Mflv_3826 | ruvC | 9e-68 | 71.01% (169) | Holliday junction resolvase |
| M. tuberculosis H37Rv | Rv2594c | ruvC | 1e-66 | 71.93% (171) | Holliday junction resolvase |
| M. leprae Br4923 | MLBr_00481 | ruvC | 1e-67 | 73.78% (164) | Holliday junction resolvase |
| M. marinum M | MMAR_2109 | ruvC | 3e-72 | 76.05% (167) | crossover junction endodeoxyribonuclease RuvC |
| M. avium 104 | MAV_3475 | ruvC | 4e-68 | 73.96% (169) | Holliday junction resolvase |
| M. smegmatis MC2 155 | MSMEG_2943 | ruvC | 2e-72 | 78.44% (167) | Holliday junction resolvase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1714 | ruvC | 2e-72 | 76.05% (167) | Holliday junction resolvase |
| M. vanbaalenii PYR-1 | Mvan_2573 | ruvC | 3e-68 | 72.19% (169) | Holliday junction resolvase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00481|M.leprae_Br4923 ----------------------------MRVMGVDPGLTRCGLSVVENGR
MAV_3475|M.avium_104 ----------------------------MRVMGVDPGLTRCGLSVVESGR
Mb2625c|M.bovis_AF2122/97 ----------------------------MRVMGVDPGLTRCGLSLIESGR
Rv2594c|M.tuberculosis_H37Rv ----------------------------MRVMGVDPGLTRCGLSLIESGR
MMAR_2109|M.marinum_M ----------------------------MRVMGVDPGLTRCGLSLIESGQ
MUL_1714|M.ulcerans_Agy99 ----------------------------MRVMGVDPGLTRCGLSLIESGQ
Mflv_3826|M.gilvum_PYR-GCK MRGARVPARSRRSQKLSSRTNVHRKGAGVRVMGVDPGLTRCGLSVIESGR
Mvan_2573|M.vanbaalenii_PYR-1 ----------------------------MRVMGVDPGLTRCGLSVIESGK
MSMEG_2943|M.smegmatis_MC2_155 ----------------------------MRVMGVDPGLTRCGLSMIESGK
MAB_2884c|M.abscessus_ATCC_199 --------------------------------------------MVEASS
::* .
MLBr_00481|M.leprae_Br4923 GSQVVALDVDVVRTPSDAPVSKRLLAVSDVVEHWLDAHHPDVMAIERVFS
MAV_3475|M.avium_104 GRTVVALDVDVVRTPSDAPLAERLLSISDAVEHWLATHQPDVVAIERVFS
Mb2625c|M.bovis_AF2122/97 GRQLTALDVDVVRTPSDAALAQRLLAISDAVEHWLDTHHPEVVAIERVFS
Rv2594c|M.tuberculosis_H37Rv GRQLTALDVDVVRTPSDAALAQRLLAISDAVEHWLDTHHPEVVAIERVFS
MMAR_2109|M.marinum_M GRQLTALDVDVVRTPSDAPLSSRLLAINEAVEHWLETHRPDVVAIERVFS
MUL_1714|M.ulcerans_Agy99 GRQLTALDVDVVRTPSDAPLSSRLLAINEAVEHWLETHRPDVVAIERVFS
Mflv_3826|M.gilvum_PYR-GCK GRQVIALDVDVVRTPSDHPLAHRLLAISNAVEHWLDTHRPDVIAIERVFS
Mvan_2573|M.vanbaalenii_PYR-1 GRQVIALDVDVVRTPAGHPLAHRLLAISDAVDHWLDTHRPDVIAIERVFS
MSMEG_2943|M.smegmatis_MC2_155 GRQVIALDVDVVRTPADTPLQKRLLTISDAAEHWMDTHRPDVIAIERVFA
MAB_2884c|M.abscessus_ATCC_199 GRKVVALDVDVVRTPANQPLPQRLLVISEVVEHWMETHRPDVVAIERVFA
* : **********:. .: *** :.:..:**: :*:*:*:******:
MLBr_00481|M.leprae_Br4923 QQNVSTVMGTAQAGGVIALAAARRGIDVHFHTPSEVKAAVTGNGAANKAQ
MAV_3475|M.avium_104 QLNVTTVMGTAQAGGVVALAAAKRGIGVHFHTPSEVKAAVTGNGAANKAQ
Mb2625c|M.bovis_AF2122/97 QLNVTTVMGTAQAGGVIALAAAKRGVDVHFHTPSEVKAAVTGNGSADKAQ
Rv2594c|M.tuberculosis_H37Rv QLNVTTVMGTAQAGGVIALAAAKRGVDVHFHTPSEVKAAVTGNGSADKAQ
MMAR_2109|M.marinum_M QQNVKTVMGTAQAGGVVALAAAKRGVEVHFHTPSEVKAAVTGNGTADKAQ
MUL_1714|M.ulcerans_Agy99 QQNVKTVMGTAQAGGVVALAAAKRGVEVHFHTPSEVKAAVTGNGTADKAQ
Mflv_3826|M.gilvum_PYR-GCK NQNANTAMGTAQAGGVVALAAARRDIDVHFHTPSEVKAAVTGNGRADKAQ
Mvan_2573|M.vanbaalenii_PYR-1 NQNANTAMGTAQAGGVIALAAARRDIDVHFHTPSEVKAAVTGNGRADKAQ
MSMEG_2943|M.smegmatis_MC2_155 NQNANTAMGTAQAGGVIALAAAKRDIEVHFHTPSEVKAAVTGSGRADKSQ
MAB_2884c|M.abscessus_ATCC_199 QQNVSTVMGTAQAGGVIALCAAKRGIEVYFHTPSEVKAAVTGNGRADKSQ
: *..*.*********:**.**:*.: *:*************.* *:*:*
MLBr_00481|M.leprae_Br4923 VTAMVTRILALQAKPTPADAADALALAICHCWRAPMIARMAEAEALGARH
MAV_3475|M.avium_104 VTAMVTRILALQAKPTPADAADALALAICHCWRAPMIARMARAEALAAQQ
Mb2625c|M.bovis_AF2122/97 VTAMVTKILALQAKPTPADAADALALAICHCWRAPTIARMAEATSRAEAR
Rv2594c|M.tuberculosis_H37Rv VTAMVTKILALQAKPTPADAADALALAICHCWRAPTIARMAEATSRAEAR
MMAR_2109|M.marinum_M VTAMVTRILALQTKPTPADAADALALAICHCWRAPMIAQMAKAHALAE--
MUL_1714|M.ulcerans_Agy99 VTAMVTRILALQTKPTPADAADALALAICHCWRAPMIAQMAKAHALAE--
Mflv_3826|M.gilvum_PYR-GCK VTEMVTRILALQQKPTPADAADALALAICHCWRAPMISRMAAAEEMAA--
Mvan_2573|M.vanbaalenii_PYR-1 VTEMVTRILALQQKPTPADAADALALAICHCWRAPMLGRMAAAEEMAA--
MSMEG_2943|M.smegmatis_MC2_155 VTEMVTRILALQAKPTPADAADALALAICHCWRAPMIARMAAAEAMAE--
MAB_2884c|M.abscessus_ATCC_199 VTAMVTRILNLQAAPKPADAADALALAICHCWRAPMLARMAKAEAVAE--
** ***:** ** *.******************* :.:** * .
MLBr_00481|M.leprae_Br4923 RQAYRAKVAGEVKATR----------
MAV_3475|M.avium_104 RQKYKDKVDATLRAAR----------
Mb2625c|M.bovis_AF2122/97 AAQQRHAYLAKLKAAR----------
Rv2594c|M.tuberculosis_H37Rv AAQQRHAYLAKLKAAR----------
MMAR_2109|M.marinum_M --QQRRSYTAKLKAAR----------
MUL_1714|M.ulcerans_Agy99 --QQRRSYTAKLKAAR----------
Mflv_3826|M.gilvum_PYR-GCK --EQRRKYQATLKAKTR--QVSRSAR
Mvan_2573|M.vanbaalenii_PYR-1 --EQRRKYQATLKAKAKAAQMSRSTR
MSMEG_2943|M.smegmatis_MC2_155 --EQRRKFQAKIKAARG---------
MAB_2884c|M.abscessus_ATCC_199 --EQRKAFAARMKAAAR---------
: . ::*