For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVLIKGVLLYGEGDQVDVLIEDGQITRIEAGIGAGIETEVDTAIEAPGQILLPGFVDLHTHLREPGREDT ETIDSGSAAAALGGYTAVFAMANTNPVADSPVVTDHVWQRGQQVGIVDVHPVGAVTVGLEGKQLTEMAMM ARGAGRVRMFSDDGKCVHDPLVMRRALEYAKGLGVLIAQHAEEPRLTVGSVAHEGANAARLGLTGWPRAA EESIVARDAILSRDTGAPVHICHASTAGTVELLKWAKGQGISITAEVTPHHLLLDDGRLSSYDGVYRVNP PLRESSDAAALRQALADGVIDCVATDHAPHADHEKCCEFAAARPGMLGLQTALSVVVQTMVRPGLLTWRD IARVMSERPAQIVGLPDQGRPIAVGEPANLTIVDPDASWTVSGDGLASKSANTPFESMTLPATVTATLLR GRITARDGRVASMSHLCDEDLAQ*
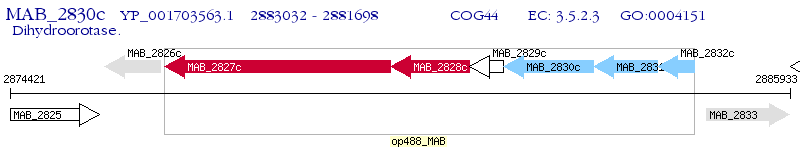
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2830c | pyrC | - | 100% (444) | dihydroorotase |
| M. abscessus ATCC 19977 | MAB_2817c | - | 2e-21 | 25.00% (456) | phenylhydantoinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1416 | pyrC | 0.0 | 80.28% (431) | dihydroorotase |
| M. gilvum PYR-GCK | Mflv_3746 | pyrC | 0.0 | 78.75% (433) | dihydroorotase |
| M. tuberculosis H37Rv | Rv1381 | pyrC | 0.0 | 80.51% (431) | dihydroorotase |
| M. leprae Br4923 | MLBr_00533 | pyrC | 0.0 | 80.65% (429) | dihydroorotase |
| M. marinum M | MMAR_2196 | pyrC | 0.0 | 79.77% (430) | dihydroorotase PyrC |
| M. avium 104 | MAV_3392 | pyrC | 0.0 | 81.48% (432) | dihydroorotase |
| M. smegmatis MC2 155 | MSMEG_3044 | pyrC | 0.0 | 80.28% (431) | dihydroorotase |
| M. thermoresistible (build 8) | TH_3497 | - | 8e-82 | 78.01% (191) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_1781 | pyrC | 0.0 | 79.30% (430) | dihydroorotase |
| M. vanbaalenii PYR-1 | Mvan_2661 | pyrC | 0.0 | 79.58% (431) | dihydroorotase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3746|M.gilvum_PYR-GCK -------MS------------VVLKSVRLYGEGDAVDVLVRDGIIAEIGT
Mvan_2661|M.vanbaalenii_PYR-1 -------MTSSSPESLRSCPPIVLKSVRLYGEGDPVDVLVRDGQIAEIGE
Mb1416|M.bovis_AF2122/97 -------MS------------VLIRGVRPYGEGERVDVLVDDGQIAQIGP
Rv1381|M.tuberculosis_H37Rv -------MS------------VLIRGVRPYGEGERVDVLVDDGQIAQIGP
MMAR_2196|M.marinum_M -------MS------------VLLRGVRLYGEGERVDVLVDDNQIADIGT
MUL_1781|M.ulcerans_Agy99 -------MS------------VLLRGVRLYGEGERVDVLVDDNQIADIGT
MLBr_00533|M.leprae_Br4923 -------MS------------VLLRGVRLYGEGEQVDVLVEGEQIANIGA
MAV_3392|M.avium_104 -------MS------------VLLRGVRLYGEGDRVDVLVDDGQIADIGA
MSMEG_3044|M.smegmatis_MC2_155 MRDHNSTAP------------VLIRGVKPYGEGDPVDVLVDDGQIARIGA
MAB_2830c|M.abscessus_ATCC_199 -------MS------------VLIKGVLLYGEGDQVDVLIEDGQITRIEA
TH_3497|M.thermoresistible__bu ----------------------LIRNVRPYGEGEPVDVLVADGQIAEIGE
:::.* ****: ****: . *: *
Mflv_3746|M.gilvum_PYR-GCK GLSA------DETIDCTGRVLLPGFVDLHTHLREPGREYAEDIETGSAAA
Mvan_2661|M.vanbaalenii_PYR-1 RLTAP---TDADSIDCEGKILLPGFVDLHTHLREPGREYAEDIETGSAAA
Mb1416|M.bovis_AF2122/97 DLAI---PDTADVIDATGHVLLPGFVDLHTHLREPGREYAEDIETGSAAA
Rv1381|M.tuberculosis_H37Rv DLAI---PDTADVIDATGHVLLPGFVDLHTHLREPGREYAEDIETGSAAA
MMAR_2196|M.marinum_M GLAI---PDTADVIDATDHVLLPGLVDLHTHLREPGREYAEDIETGSAAA
MUL_1781|M.ulcerans_Agy99 GLAI---PDTADVIDATDHVLLPGLVDLHTHLREPGREYAEDIETGSAAA
MLBr_00533|M.leprae_Br4923 GIDI---PDTADVIDAIDQVLLPGLVDLHTHLREPGREYAEDIETGSAAA
MAV_3392|M.avium_104 GLDI---PDTADVIDATGQVLLPGFVDLHTHLREPGREYAEDIETGSAAA
MSMEG_3044|M.smegmatis_MC2_155 DLPV---PETADVIDARGQVLLPGFVDLHTHLREPGREYAEDIETGSAAA
MAB_2830c|M.abscessus_ATCC_199 GIGAGIETEVDTAIEAPGQILLPGFVDLHTHLREPGREDTETIDSGSAAA
TH_3497|M.thermoresistible__bu KLPIPGDGELGDVIDADGQILLPGFVDLHTHLREPGREYAEDIETGSAAA
: *:. .::****:************* :* *::*****
Mflv_3746|M.gilvum_PYR-GCK ALGGYTAVFAMANTDPVADSVVVTDHVWQRGQQVGLVDVHPVGAVTVGLA
Mvan_2661|M.vanbaalenii_PYR-1 ALGGYTAVFAMANTDPVADSAVVTDHVWHRGQQVGLVDVHPVGAVTVGLD
Mb1416|M.bovis_AF2122/97 ALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVGLA
Rv1381|M.tuberculosis_H37Rv ALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVGLA
MMAR_2196|M.marinum_M ALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVGLA
MUL_1781|M.ulcerans_Agy99 ALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTIGLA
MLBr_00533|M.leprae_Br4923 ALGGYTAVFAMPNTTPVADSPVVTDHVWRRGQQVGLVDVHPVGAVTVGLA
MAV_3392|M.avium_104 ALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTMGLA
MSMEG_3044|M.smegmatis_MC2_155 ALGGYTAVFAMANTDPVADTVVVTDHVWRRGQEVGLVDVHPVGAITMGLK
MAB_2830c|M.abscessus_ATCC_199 ALGGYTAVFAMANTNPVADSPVVTDHVWQRGQQVGIVDVHPVGAVTVGLE
TH_3497|M.thermoresistible__bu ALGGYTAVFAMANTDPVADSPVVTDHVWRRGQQVGLVDVHPVGAVTVGLE
***********.** ****: *******:***:**:********:*:**
Mflv_3746|M.gilvum_PYR-GCK GKQLTEMGLMAAGTGRVRMFSDDGVCVDDPLLMRRALEYASGLGVLIAQH
Mvan_2661|M.vanbaalenii_PYR-1 GKQLTEMGLMAAGAGQVRMFSDDGVCVDDPLVMRRALEYASGLGVLIAQH
Mb1416|M.bovis_AF2122/97 GAELTEMGMMNAGAAQVRMFSDDGVCVHDPLIMRRALEYATGLGVLIAQH
Rv1381|M.tuberculosis_H37Rv GAELTEMGMMNAGAAQVRMFSDDGVCVHDPLIMRRALEYATGLGVLIAQH
MMAR_2196|M.marinum_M GTELTEMGMMNAGVAAVRMFSDDGICVHDPLIMRRALEYATGLGVLIAQH
MUL_1781|M.ulcerans_Agy99 GTELTEMGMMNAGVAAVRMFSDDGICVHDPLIMRRALEYATGLGVLIAQH
MLBr_00533|M.leprae_Br4923 GAELTEMGMMAAGAAQVRIFSDDGNCVHNPLVMRRALEYATGLGVLIAQH
MAV_3392|M.avium_104 GKELTEMGMMAAGAAQVRMFSDDGVCVHDPLVMRRALEYATGLGVLIAQH
MSMEG_3044|M.smegmatis_MC2_155 GAQLTEMGLMAAGAGQVRMFSDDGVCVENPLVMRRALEYATGLGVLIAQH
MAB_2830c|M.abscessus_ATCC_199 GKQLTEMAMMARGAGRVRMFSDDGKCVHDPLVMRRALEYAKGLGVLIAQH
TH_3497|M.thermoresistible__bu GRQLTEMGLMAAGAARVRMFSDDGLCVHDPLVMRRALEYARGLGVLIAQH
* :****.:* *.. **:***** **.:**:******** *********
Mflv_3746|M.gilvum_PYR-GCK AEEPRLTVGAVAHEG-----PNAARLGLAGWPRAAEESIVARDALLARDA
Mvan_2661|M.vanbaalenii_PYR-1 AEEPRLTVGAVAHEG-----PNAARLGLAGWPRAAEESIVARDALLARDA
Mb1416|M.bovis_AF2122/97 AEEPRLTVGAFAHEG-----PMAARLGLAGWPRAAEESIVARDALLARDA
Rv1381|M.tuberculosis_H37Rv AEEPRLTVGAVAHEG-----PMAARLGLAGWPRAAEESIVARDALLARDA
MMAR_2196|M.marinum_M AEEPRLTVGAVAHEG-----PNAAQLGLAGWPRAAEESIVARDALLARDA
MUL_1781|M.ulcerans_Agy99 AEEPRLTVGAVAHEG-----PSAAQLGLAGWPRAAEESIVARDALLTRDA
MLBr_00533|M.leprae_Br4923 AEEPRLTVGAVAHEG-----PTAARLGLSGWPRAAEESIVARDALLARDA
MAV_3392|M.avium_104 AEEPRLTVGAVAHEG-----PTAARLGLAGWPRAAEESIVARDALLARDA
MSMEG_3044|M.smegmatis_MC2_155 AEEPRLTVGAVAHEG-----PIAARLGLAGWPRAAEESIVARDALLARDA
MAB_2830c|M.abscessus_ATCC_199 AEEPRLTVGSVAHEG-----ANAARLGLTGWPRAAEESIVARDAILSRDT
TH_3497|M.thermoresistible__bu AEEPRLTVGAVAHXXXWPACPARCRPGYRSPPTWCASAPPGRPNAVRRNR
*********:.** . .: * . * . .: .* : *:
Mflv_3746|M.gilvum_PYR-GCK GARVHICHASTAGTVELVRWAKEQGIAITAEVTP-----HHLLLDDT---
Mvan_2661|M.vanbaalenii_PYR-1 GARVHICHASTAGTVELLKWAKSQGISITAEVTP-----HHLLLDDS---
Mb1416|M.bovis_AF2122/97 GARVHICHASAAGTVEILKWAKDQGISITAEVTP-----HHLLLDDA---
Rv1381|M.tuberculosis_H37Rv GARVHICHASAAGTVEILKWAKDQGISITAEVTP-----HHLLLDDA---
MMAR_2196|M.marinum_M GARVHICHASTVGTVEILKWAKSQGISITAEVTP-----HHLLLDDR---
MUL_1781|M.ulcerans_Agy99 GARVHICHASTVGTVEILKWAKSQGISITAEVTP-----HHLLLDDR---
MLBr_00533|M.leprae_Br4923 GARVHICHASTAGTVEILKWAKEQGISITAEVTP-----HHLLLDDS---
MAV_3392|M.avium_104 GARVHICHASTAGTVEIVKWAKQQGISITAEVTP-----HHLMLDDS---
MSMEG_3044|M.smegmatis_MC2_155 GARVHICHASTAGTVELLKWAKAQGISITAEVTP-----HHLLLDDS---
MAB_2830c|M.abscessus_ATCC_199 GAPVHICHASTAGTVELLKWAKGQGISITAEVTP-----HHLLLDDG---
TH_3497|M.thermoresistible__bu SSPRRIGPTPPPGTSRPATDVGRLGSADRARRPGPRCRGAWWLGHDAPVT
.: :* :.. ** . . * : *. . : .*
Mflv_3746|M.gilvum_PYR-GCK RLETYD-----GRNRVNPPLREVSDTLALRQALADGVID----CVATDHA
Mvan_2661|M.vanbaalenii_PYR-1 RLDTYD-----GVNRVNPPLREVSDAVALRRALADGVID----CVATDHA
Mb1416|M.bovis_AF2122/97 RLASYD-----GVNRVNPPLREASDAVALRQALADGIID----CVATDHA
Rv1381|M.tuberculosis_H37Rv RLASYD-----GVNRVNPPLREASDAVALRQALADGIID----CVATDHA
MMAR_2196|M.marinum_M RLSGYD-----GVNRVNPPLREASDAMALRQALADGIID----CVATDHA
MUL_1781|M.ulcerans_Agy99 RLSGYD-----GVNRVNPPLREASDAMALRQALADGIID----CVATDHA
MLBr_00533|M.leprae_Br4923 RLASYD-----GVNRVNPPLRETADAVALRQALADGIID----CVTTDHA
MAV_3392|M.avium_104 RLASYD-----GRNRVNPPLREAADAAALRRALADGVID----CVATDHA
MSMEG_3044|M.smegmatis_MC2_155 RLETYD-----GRNRVNPPLREAEDAAALRQALADGVID----CVATDHA
MAB_2830c|M.abscessus_ATCC_199 RLSSYD-----GVYRVNPPLRESSDAAALRQALADGVID----CVATDHA
TH_3497|M.thermoresistible__bu ELTLYDVLTTLDVAQVDDDLFEATQLDNPAHHIVGGHIAGQAVVAASRTA
.* ** . :*: * * : : :..* * .:: *
Mflv_3746|M.gilvum_PYR-GCK PH-GEHEKMCEFANARPGMLGLQT---------ALSVVVETMVQPG----
Mvan_2661|M.vanbaalenii_PYR-1 PH-GAHEKCCEFANARPGMLGLQT---------ALSVVVETLVNPG----
Mb1416|M.bovis_AF2122/97 PH-AEHEKCVEFAAARPGMLGLQT---------ALSVVVQTMVAPG----
Rv1381|M.tuberculosis_H37Rv PH-AEHEKCVEFAAARPGMLGLQT---------ALSVVVQTMVAPG----
MMAR_2196|M.marinum_M PH-AEHEKCVEFSAARPGMLGLQT---------ALSVVVQTMVQPG----
MUL_1781|M.ulcerans_Agy99 PH-AEHEKCVEFSAARPGMLGLQT---------ALSVVVQTMVQPG----
MLBr_00533|M.leprae_Br4923 PH-ADHEKCVEFSAARPGMLGLQT---------ALSVVVHTMVVPG----
MAV_3392|M.avium_104 PH-AEHEKCVEFSAARPGMLGLQT---------ALSIVVHTMVRPG----
MSMEG_3044|M.smegmatis_MC2_155 PH-AEHEKCCEFSVARPGMLGLQT---------ALSVVVETMVRPG----
MAB_2830c|M.abscessus_ATCC_199 PH-ADHEKCCEFAAARPGMLGLQT---------ALSVVVQTMVRPG----
TH_3497|M.thermoresistible__bu PQRTAHSAHVYFLRAGDARYPVQMRVDRLRDGGSLSTXRVTATQDGQVLL
*: *. * * . :* :** * . *
Mflv_3746|M.gilvum_PYR-GCK --------PLTWRDVARVMSEAP---------ARIVGLPDQGRPLEVG-E
Mvan_2661|M.vanbaalenii_PYR-1 --------LLTWRDVARVMSEAP---------ARIVGLPDQGRPLEVG-E
Mb1416|M.bovis_AF2122/97 --------LLSWRDIARVMSENP---------ACIARLPDQGRPLEVG-E
Rv1381|M.tuberculosis_H37Rv --------LLSWRDIARVMSENP---------ACIARLPDQGRPLEVG-E
MMAR_2196|M.marinum_M --------LLSWRDVARVMSENP---------ARIVGLPDHGRPLEVG-E
MUL_1781|M.ulcerans_Agy99 --------LLSWRDVARVMSENP---------ARIVGLPDHGRPLEVG-E
MLBr_00533|M.leprae_Br4923 --------LLSWRDVAQVMSENP---------ARIAGLPDHGRPLEVG-E
MAV_3392|M.avium_104 --------LLTWRDVAKVMSENP---------ARIANLPDHGRPLEVG-E
MSMEG_3044|M.smegmatis_MC2_155 --------LLTWRGVAKVMSEAP---------AAIVGLPDQGRPLEVG-E
MAB_2830c|M.abscessus_ATCC_199 --------LLTWRDIARVMSERP---------AQIVGLPDQGRPIAVG-E
TH_3497|M.thermoresistible__bu EALASFTVRIDSVDTHHPMPDVPDPDTLPPVTEQLVAYADEFGGLWVRPQ
: . : *.: * :. .*. : * :
Mflv_3746|M.gilvum_PYR-GCK PANLTVVDPDARWTVSGPALASRS----------DNTPYEAMELPGVVT-
Mvan_2661|M.vanbaalenii_PYR-1 PANLTVVDPAARWTVKGPELASRS----------DNTPYEDMELPAVVT-
Mb1416|M.bovis_AF2122/97 PANLTVVDPDATWTVTGADLASRS----------ANTPFESMSLPATVT-
Rv1381|M.tuberculosis_H37Rv PANLTVVDPDATWTVTGADLASRS----------ANTPFESMSLPATVT-
MMAR_2196|M.marinum_M PANLTVIDPDATWTVTGADLASRS----------ANTPYESMTLPARVT-
MUL_1781|M.ulcerans_Agy99 PANLTVIDPDATWTVTGADLASRS----------ANTPYESMTLPARVT-
MLBr_00533|M.leprae_Br4923 PANLTVVDPDVTWTVTGDGLASRS----------ANTPYEAMTLPATVT-
MAV_3392|M.avium_104 PANLTVVDPDASWTVAGSDLASRS----------ANTPYEAMTLPATVT-
MSMEG_3044|M.smegmatis_MC2_155 PANLTVVDPDATWTVAGTELASRS----------DNTPFETMTLPATVT-
MAB_2830c|M.abscessus_ATCC_199 PANLTIVDPDASWTVSGDGLASKS----------ANTPFESMTLPATVT-
TH_3497|M.thermoresistible__bu PIERRYIDPPPRVAYDLPEPPARTRMWWRATEPVPDDPALNSALLTYVTG
* : :** : .::: : * * **
Mflv_3746|M.gilvum_PYR-GCK -ATLLRGTVTARDGKAGA--------------------------------
Mvan_2661|M.vanbaalenii_PYR-1 -ATLLRGKVTARDGKVGA--------------------------------
Mb1416|M.bovis_AF2122/97 -ATLLRGKVTARDGKIRA--------------------------------
Rv1381|M.tuberculosis_H37Rv -ATLLRGKVTARDGKIRA--------------------------------
MMAR_2196|M.marinum_M -ATLLRGKITARDGKSPA--------------------------------
MUL_1781|M.ulcerans_Agy99 -ATLLRGKITARDGKSPA--------------------------------
MLBr_00533|M.leprae_Br4923 -ATLLRGKVTARGQKSPV--------------------------------
MAV_3392|M.avium_104 -ATLLRGKVTARDGKCPV--------------------------------
MSMEG_3044|M.smegmatis_MC2_155 -ATLLRGRVTARDGKSAAAGGGSVQ-------------------------
MAB_2830c|M.abscessus_ATCC_199 -ATLLRGRITARDGRVASMSHLCDEDLAQ---------------------
TH_3497|M.thermoresistible__bu TSTLEAAMVAHRTTPLHSFNALIDHALWFHRPAHLTDWVLADQFSPSSLG
:** . :: *
Mflv_3746|M.gilvum_PYR-GCK ------------------------------------
Mvan_2661|M.vanbaalenii_PYR-1 ------------------------------------
Mb1416|M.bovis_AF2122/97 ------------------------------------
Rv1381|M.tuberculosis_H37Rv ------------------------------------
MMAR_2196|M.marinum_M ------------------------------------
MUL_1781|M.ulcerans_Agy99 ------------------------------------
MLBr_00533|M.leprae_Br4923 ------------------------------------
MAV_3392|M.avium_104 ------------------------------------
MSMEG_3044|M.smegmatis_MC2_155 ------------------------------------
MAB_2830c|M.abscessus_ATCC_199 ------------------------------------
TH_3497|M.thermoresistible__bu GRGLATATMYNRTGELVCTATQELYFGRPARSGSRR