For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNLTDSTPGGSNLAGAVEGTGINKGELFSSYDVDAFEVPGGRDELWRFTPLRRLRGLHDGTAVATAAAPV AVNASGGVTVETVERSDDRLGQGGIPSDRVAAQAYSSFSTATVVSVGKQQVVADPIEISLSGPGQGQVAY GHTQVRAGALSESAVVIDYRGSGTYAENVEFVLDDAARLTVVSIADWADDAVHVTTHHAKLGKDAVLRHI AVTLGGDVVRLTGTVRFGAPGGDAELLGLYYADEGQFFEHRLLVDHAQPNCKSNVVYKGALQGDPSSDKP DAHTVWIGDVLIRAEATGTDTFELNRNLILTDGARADSVPNLEIETGEIAGAGHASATGRFDDEQLFYLR SRGIPEEDARRLVVRGFFQDIIGRIGIESVRDRLTEAIEQELQANAN*
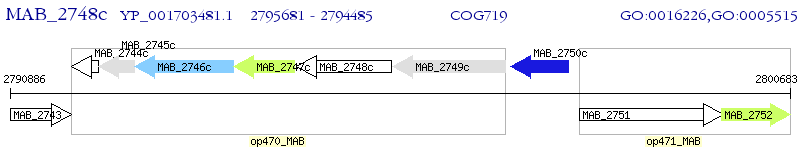
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2748c | - | - | 100% (398) | hypothetical protein MAB_2748c |
| M. abscessus ATCC 19977 | MAB_2749c | - | 4e-17 | 24.26% (235) | putative FeS assembly protein SufB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1497 | - | 1e-162 | 71.92% (381) | hypothetical protein Mb1497 |
| M. gilvum PYR-GCK | Mflv_3676 | - | 1e-160 | 69.70% (396) | FeS assembly protein SufD |
| M. tuberculosis H37Rv | Rv1462 | - | 1e-161 | 71.65% (381) | hypothetical protein Rv1462 |
| M. leprae Br4923 | MLBr_00594 | - | 1e-158 | 71.05% (380) | hypothetical protein MLBr_00594 |
| M. marinum M | MMAR_2267 | - | 1e-164 | 73.39% (372) | hypothetical protein MMAR_2267 |
| M. avium 104 | MAV_3317 | sufD | 1e-167 | 73.51% (385) | FeS assembly protein SufD |
| M. smegmatis MC2 155 | MSMEG_3123 | sufD | 1e-168 | 72.54% (397) | FeS assembly protein SufD |
| M. thermoresistible (build 8) | TH_1610 | - | 1e-106 | 72.98% (248) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1863 | - | 1e-164 | 73.39% (372) | hypothetical protein MUL_1863 |
| M. vanbaalenii PYR-1 | Mvan_2734 | - | 1e-165 | 72.49% (389) | FeS assembly protein SufD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1497|M.bovis_AF2122/97 MTAPGLTAAVEG----------IAHNKGELFASFDVDAFEVPHGRDEIWR
Rv1462|M.tuberculosis_H37Rv MTAPGLTAAVEG----------IAHNKGELFASFDVDAFEVPHGRDEIWR
MMAR_2267|M.marinum_M MTVDNLTTAAA-------------LNKGELFASFDVDAFEVPHGRDELWR
MUL_1863|M.ulcerans_Agy99 MTVDNLTTAAA-------------LNKGELFASFDVDAFEVPHGRDELWR
MAV_3317|M.avium_104 --MTNLTEAVEGSS-------LAAANKGELFASFDVDAFEVPHGRDEIWR
MLBr_00594|M.leprae_Br4923 ------MTAVEGSS-------LTALNKGRLFASFDVDAFEVPGGRDEIWR
Mflv_3676|M.gilvum_PYR-GCK -MSRDLTSAVEGSVKSG---SVLNLNKGELFSSFDVNAFEVPGGRDELWR
Mvan_2734|M.vanbaalenii_PYR-1 -MASQLTQASEGTAKSG---SVLNLNKGELFSSFDVSAFEVPGGRDEIWR
MSMEG_3123|M.smegmatis_MC2_155 -MASQLTQASEGSAKSG---SVLNLNKGEVFTSFDVNAFEVPHGRDEIWR
TH_1610|M.thermoresistible__bu --------------------------------------------------
MAB_2748c|M.abscessus_ATCC_199 --MSNLTDSTPGGSNLAGAVEGTGINKGELFSSYDVDAFEVPGGRDELWR
Mb1497|M.bovis_AF2122/97 FTPLRRLRGLHDGSARATGSAT----ITVSERPGVYTQTVRRGDPRLGEG
Rv1462|M.tuberculosis_H37Rv FTPLRRLRGLHDGSARATGSAT----ITVSERPGVYTQTVRRGDPRLGEG
MMAR_2267|M.marinum_M FTPLKRLRGLHDGSAAASGSAQ----LTVGEQAGLRVETVRRGDERLGQG
MUL_1863|M.ulcerans_Agy99 FTPLKRLRGLHDGSAAASGSAQ----LTVGEQAGLRVETVRRGDERLGQG
MAV_3317|M.avium_104 FTPLRRLRGLHDGSAPATGKAE----VSVGEQPGLRVQTVRRGDERLGQG
MLBr_00594|M.leprae_Br4923 FTPLRRLRGLHDGSAVANGKVH----IRVSQQSGVQTEIVGRGDGRLGQG
Mflv_3676|M.gilvum_PYR-GCK FTPLKRLRGLHDGSATPSGTAG----ITVTEREGVTVETVARGDARLGQA
Mvan_2734|M.vanbaalenii_PYR-1 FTPLKRLRGLHDGSAPATGTAT----IEVTTPDDVWVDKTTRDDGRLGQA
MSMEG_3123|M.smegmatis_MC2_155 FTPLRRLRGLHNGTAGADGKATGSALIEVSERPGVTIETVERGDERLGQG
TH_1610|M.thermoresistible__bu --------------------------------------------------
MAB_2748c|M.abscessus_ATCC_199 FTPLRRLRGLHDGTAVATAAAP----VAVNASGGVTVETVERSDDRLGQG
Mb1497|M.bovis_AF2122/97 GVPTDRVAAQAFSSFNSATLVTVERDTQVVEPVGITVTGPGEGAVAYGHL
Rv1462|M.tuberculosis_H37Rv GVPTDRVAAQAFSSFNSATLVTVERDTQVVEPVGITVTGPGEGAVAYGHL
MMAR_2267|M.marinum_M GVPADRVAAQAFSSFNSATLVTVGRDTQLGEPVNITITGPGEGAVAYGHL
MUL_1863|M.ulcerans_Agy99 GVPADRVAAQAFSSFNSATLVTVGRDTQLGEPVNITITGPGKGAVAYGHL
MAV_3317|M.avium_104 GIPTDRVAAQAFSSFNSATLITVQRDTQIAEPIGVTVTGPGEGAVAYGHL
MLBr_00594|M.leprae_Br4923 GIPTDRVAAQAFSSFQSATVVTVGRDMQIATPINIAVTGPDKGAVVYGHL
Mflv_3676|M.gilvum_PYR-GCK GVPSDRVAAQAFSSFESATVVTVARDTEVAEPIEIVIEGPGEYAVAYGHL
Mvan_2734|M.vanbaalenii_PYR-1 GAPADRVAAQAYSSFESATLIEIPNQAVIRDPIEVRITGPGEGATAYGHL
MSMEG_3123|M.smegmatis_MC2_155 GVPADRVAAQAFSSFSNATVVTVGRNVQVDEPVEITITGPGEGNTAYGHV
TH_1610|M.thermoresistible__bu --------------------------------------------------
MAB_2748c|M.abscessus_ATCC_199 GIPSDRVAAQAYSSFSTATVVSVGKQQVVADPIEISLSGPGQGQVAYGHT
Mb1497|M.bovis_AF2122/97 QVRIEELGEAVVVIDHRGGGTYADNVEFVVDDAARLTAVWIADWADDTVH
Rv1462|M.tuberculosis_H37Rv QVRIEELGEAVVVIDHRGGGTYADNVEFVVDDAARLTAVWIADWADNTVH
MMAR_2267|M.marinum_M QIRVEELGEAVVVVDHRGSGTYADNVEFVVGDCARLTVVWIADWADDTVH
MUL_1863|M.ulcerans_Agy99 QIRVEELGEAVVVVDHRGSGTYADNVEFVVGDCARLTVVWIADWADDTVH
MAV_3317|M.avium_104 QIRVEELAEAVVVIDHRGSGTLADNVEFVVDDAARLTVVWLADWADDAVH
MLBr_00594|M.leprae_Br4923 QIRVCKFGEAVVVIDHRGSGTYADNVEFIVEAAARLTVVWIADWADDMVH
Mflv_3676|M.gilvum_PYR-GCK QIRVEELSRAIVVVDLRGSGVYADNVEIIVGDSAGLGLIWIADWADDTVH
Mvan_2734|M.vanbaalenii_PYR-1 QIRVGELAEAVVVIDHRGSGTYADNIEFVVGDGATLRVVSIADWADDVVH
MSMEG_3123|M.smegmatis_MC2_155 QVRVEELGEAVVVIDHRGSGTYADNIEFVVGDAATLKVVSIADWADDTVH
TH_1610|M.thermoresistible__bu -VRVEQLADAVVVIDQRGSGVYADNIEFVVGDGATLKVVSIADWAGDAVN
MAB_2748c|M.abscessus_ATCC_199 QVRAGALSESAVVIDYRGSGTYAENVEFVLDDAARLTVVSIADWADDAVH
:* :. : **:* **.*. *:*:*::: * * : :****.: *:
Mb1497|M.bovis_AF2122/97 LSAHHARIGKDAVLRHVTVMLGGDVVRMSAGVRFCGAGGDAELLGLYFAD
Rv1462|M.tuberculosis_H37Rv LSAHHARIGKDAVLRHVTVMLGGDVVRMSAGVRFCGAGGDAELLGLYFAD
MMAR_2267|M.marinum_M LSAHHAKLGKDAVLRHVTVTLGGEVVRMSANVRFTAPGGDADLLGLYFAD
MUL_1863|M.ulcerans_Agy99 LSAHHAKLGKDAVLRHVTVTLGGEVVRMSANVRFTAPGGDADLLGLYFAD
MAV_3317|M.avium_104 VSMHHARLGKDAVLRHVAVTLGGEVVRLTANVRYTSTGGDAELLGLYFAD
MLBr_00594|M.leprae_Br4923 LSAHHALLGKDAVLRHITVTLGGEVVRVSANVRFSGPGGDAELLGLYFAD
Mflv_3676|M.gilvum_PYR-GCK VSSHHAKLGRDAVLGHVNVTLGGDVVRTTATVRYTGPGGEAKMLGTYFAD
Mvan_2734|M.vanbaalenii_PYR-1 VTMHHALLGRDAVLRHAAVTLGGSVVRMTGRVRYTAPGGDAELLGLYYAD
MSMEG_3123|M.smegmatis_MC2_155 VSTHHATLGKDAVLRHFAVTLGGDVARLTGRVRFAAPGGDAELLGLYFAD
TH_1610|M.thermoresistible__bu VSMHHAALGKDAVLRHAAVLLGGEVVRMTGRVRFNAPGGDAELYGLYFAD
MAB_2748c|M.abscessus_ATCC_199 VTTHHAKLGKDAVLRHIAVTLGGDVVRLTGTVRFGAPGGDAELLGLYYAD
:: *** :*:**** * * ***.*.* :. **: ..**:*.: * *:**
Mb1497|M.bovis_AF2122/97 DGQHLESRLLVDHAHPDCKSNVLYKGALQGDPASSLPDAHTVWVGDVLIR
Rv1462|M.tuberculosis_H37Rv DGQHLESRLLVDHAHPDCKSNVLYKGALQGDPASSLPDAHTVWVGDVLIR
MMAR_2267|M.marinum_M DGQHLESRLLVDHAQPDCKSNVLYKGALQGDPGSALPDAHTVWVGDVLIR
MUL_1863|M.ulcerans_Agy99 DGQHLESRLLVDHAQPDCKSNVLYKGALQGDPGSALPDAHTVWVGDVLIR
MAV_3317|M.avium_104 DGQHLESRLLVDHAQPDCKSNVLYKGALQGDPASALPDAHTVWVGDVLIR
MLBr_00594|M.leprae_Br4923 DGQHLESRLLVDHAHPDCKSNVLYKGALQGDPVSSRPDAHTVWVGDVLIH
Mflv_3676|M.gilvum_PYR-GCK DGQHFESRLLVDHSQPNCKSDVLYKGALQGDPDSRKPDAHTVWVGDVLIR
Mvan_2734|M.vanbaalenii_PYR-1 AGQHFESRLLVDHSQPNCKSDVKYKGALQGDPDSRKPDAHTVWVGDVLIR
MSMEG_3123|M.smegmatis_MC2_155 DGQHFESRLLVDHAQPHCRSHVTYKGALQGDPASDKPDTHTVWVGDVLIR
TH_1610|M.thermoresistible__bu DGQHLESRLLIDHAVPHCRSNVIYKGALQSDPDSKRPDAHAVWVGDVLIR
MAB_2748c|M.abscessus_ATCC_199 EGQFFEHRLLVDHAQPNCKSNVVYKGALQGDPSSDKPDAHTVWIGDVLIR
**.:* ***:**: *.*:*.* ******.** * **:*:**:*****:
Mb1497|M.bovis_AF2122/97 AQATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
Rv1462|M.tuberculosis_H37Rv AQATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
MMAR_2267|M.marinum_M AAATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIAGAGHASATGRFDD
MUL_1863|M.ulcerans_Agy99 AAATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIAGAGHASATGRFDD
MAV_3317|M.avium_104 AEATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIAGAGHASATGRFDD
MLBr_00594|M.leprae_Br4923 PEATGTDTFEVNRNLVLTNGVRADSVPNLEIETDEIVGAGHASATGRFDD
Mflv_3676|M.gilvum_PYR-GCK AEATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
Mvan_2734|M.vanbaalenii_PYR-1 AEATGTDTFEVNRNLVLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
MSMEG_3123|M.smegmatis_MC2_155 AEATGTDTYEVNRNLVLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
TH_1610|M.thermoresistible__bu AAATGTDTYEINRNLLLTDGARADSVPNLEIETGEIVGAGHASATGRFDD
MAB_2748c|M.abscessus_ATCC_199 AEATGTDTFELNRNLILTDGARADSVPNLEIETGEIAGAGHASATGRFDD
. ******:*:****:**:*.************.**.*************
Mb1497|M.bovis_AF2122/97 EQLFYLRSRGIPEAQARRLVVRGFFGEIIAKIAVPEVRERLTAAIEHELE
Rv1462|M.tuberculosis_H37Rv EQLFYLRSRGIPEAQARRLVVRGFFGEIIAKIAVPEVRERLTAAIEHELE
MMAR_2267|M.marinum_M EQLFYLRSRGIPEEQARRLVIRGFFGEIISKIAVPEVRERLTGAIEHELE
MUL_1863|M.ulcerans_Agy99 EQLFYLRSRGIPEEQARRLVIRGFFGEIISKIAVPEVRERLTGAIEHELE
MAV_3317|M.avium_104 EQLFYLRSRGIPEEQARRLVIRGFFAEIISKIAVPEIRERLTAAIEHELA
MLBr_00594|M.leprae_Br4923 EQLFYLRSRGIGEEQARRLLVRGFFGEIISKIAVPQVRERLIAAIEHELT
Mflv_3676|M.gilvum_PYR-GCK EQLFYLRARGIPEAQARRLVVRGFFNEIIAKIAVPAVRERLTDAIERELA
Mvan_2734|M.vanbaalenii_PYR-1 EQLFYLRARGIPENQARRLVVRGFFNEIIAKIAVPAVRERLTEAIERELA
MSMEG_3123|M.smegmatis_MC2_155 EQLFYLRARGIPEDQARRLVVRGFFGELIQKIAVPAVRDRLTAAIEHELE
TH_1610|M.thermoresistible__bu EQVFYLQARGIPEDQARKLIVRGFFNEVIQKITVPEVRDRLTDAIEHELA
MAB_2748c|M.abscessus_ATCC_199 EQLFYLRSRGIPEEDARRLVVRGFFQDIIGRIGIESVRDRLTEAIEQELQ
**:***::*** * :**:*::**** ::* :* : :*:** ***:**
Mb1497|M.bovis_AF2122/97 ITES-----TEKTTVS
Rv1462|M.tuberculosis_H37Rv ITES-----TEKTTVS
MMAR_2267|M.marinum_M ITESQATEKTENTAAS
MUL_1863|M.ulcerans_Agy99 ITESQATEKTENTAAS
MAV_3317|M.avium_104 FTES-------RATAS
MLBr_00594|M.leprae_Br4923 ITES-------RSTAS
Mflv_3676|M.gilvum_PYR-GCK ITEA---------SAN
Mvan_2734|M.vanbaalenii_PYR-1 LTEG---------NNQ
MSMEG_3123|M.smegmatis_MC2_155 LTEG---------IKH
TH_1610|M.thermoresistible__bu ITEG---------KLS
MAB_2748c|M.abscessus_ATCC_199 ANAN------------
.