For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPRTSDPLRHWLFALHVRPARLAMAVMFGVLALGSALGLAGLSAWLITRAWQMPPVLDLSVAVVAVRALG ISRGIWRYCERLAGHDIALRGAATVRTHVYARLTGGPAGRLRKGAVLERIGTDIDELAETVVRSVIPAAV AAVLGIAATAVVALISIPAAAVMACALLVADVLAPALAARAARARETRGARARALHSETAVEILDHAPEL RVGGLLPHQLEVARHRRGEWAAAQDRAAAPAAWSAAAPTIAVGMASVGALAAAVELANHVAPTTLAILVL LPLAAFEASTALPAAAVGIARSRASARHLQELAPDGRGAEPTDVMPSAAAGAAVECSELHWHTGNSVSGP LSFRLPAGARMAVTGASGIGKTSLLTTLAGLTRPVSGTIRVRNDALADLPAAELPGLIRFFAEDAHLFAT TVRDNLLVARGDAPDGDLVDALSVVGLGPWLESLPQGLDTVMTAGAGSVSGGQRRRILLARALLSRVPVL LLDEPTEHLDGRDSDSIMRKLLNANSSLLAGRTVVVATHHLPDSVSCLRIQLGDTVQV
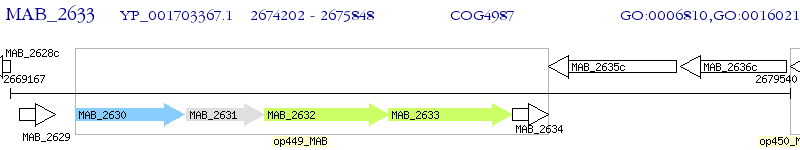
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2633 | - | - | 100% (548) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2261c | - | 2e-26 | 25.88% (483) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2632 | - | 3e-24 | 26.67% (555) | ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1646c | cydC | 1e-144 | 53.40% (530) | cytochrome' transport transmembrane ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_3588 | - | 1e-126 | 50.67% (523) | ABC transporter, transmembrane region, type 1 |
| M. tuberculosis H37Rv | Rv1620c | cydC | 1e-144 | 53.40% (530) | cytochrome' transport transmembrane ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_01113 | - | 7e-22 | 23.42% (521) | putative ABC transporter, ATP-binding component |
| M. marinum M | MMAR_2423 | cydC | 1e-143 | 51.53% (555) | cytochrome transmembrane ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3167 | - | 1e-145 | 53.27% (535) | ABC transporter, ATP-binding protein CydC |
| M. smegmatis MC2 155 | MSMEG_5661 | - | 8e-30 | 26.66% (574) | ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_4206 | - | 4e-70 | 53.56% (267) | PUTATIVE ABC transporter, transmembrane region, type 1 |
| M. ulcerans Agy99 | MUL_1601 | cydC | 1e-142 | 51.74% (545) | component linked with the assembly of cytochrome transport |
| M. vanbaalenii PYR-1 | Mvan_2827 | - | 1e-132 | 50.47% (533) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1646c|M.bovis_AF2122/97 ---------------MNRPS--AVSRRQRDLLAASGLLGPRLPRILAAVA
Rv1620c|M.tuberculosis_H37Rv ---------------MNRPS--AVSRRQRDLLAASGLLGPRLPRILAAVA
MAV_3167|M.avium_104 ---------------MPRPDSFASASSGRPLLAAAGLLSPRLPRLAAAIT
MMAR_2423|M.marinum_M ------------------------MSRSDSLSAAYDLLRPRLPRVLAAIA
MUL_1601|M.ulcerans_Agy99 -------------------------------------MRPRLPRVLAAIA
Mflv_3588|M.gilvum_PYR-GCK ----------------------MPVSRREPLLAAWPLLRPRLPRVALSVL
Mvan_2827|M.vanbaalenii_PYR-1 ------------------------MSRRDPLARAWPLLGSRAPRIALAVM
TH_4206|M.thermoresistible__bu ---------------------MRHSPTADPLWSMIPLLRPRLPRLALAVT
MAB_2633|M.abscessus_ATCC_1997 -----------------------MPRTSDPLRHWLFALHVRPARLAMAVM
MSMEG_5661|M.smegmatis_MC2_155 MSIETVARQTLYRQTHAKGGDLHALANRRLLARIWRFAARHHRRLAVFFA
MLBr_01113|M.leprae_Br4923 -------------MTMATAPATRSCDFYCTIRRLLKQLVPQRRLSLTVIT
: .
Mb1646c|M.bovis_AF2122/97 LGVLSLGSALALAGVSAWLITRAWQMPPVLDLSVAVVAVRAFAISRG---
Rv1620c|M.tuberculosis_H37Rv LGVLSLGSALALAGVSAWLITRAWQMPPVLDLSVAVVAVRAFAISRG---
MAV_3167|M.avium_104 LGVLSLGSALALAGVSAWLITRAWQMPPVLDLSVAAVAVRMFAISRG---
MMAR_2423|M.marinum_M LGVLSLGSALALAGVSAWLITRASQMPPVLDLSIAVVAVRTFAISRG---
MUL_1601|M.ulcerans_Agy99 LGVLPLGSALALAGVSAWLITRASQMPPVLDLSIAVVAVRTFAISRG---
Mflv_3588|M.gilvum_PYR-GCK LGALSLGSALALTAVSAWLITRAWQMPPVLHLTVAAVTVRALGISRG---
Mvan_2827|M.vanbaalenii_PYR-1 FGVLALGSALGLTAVSAWLITRAWEMPPVLHLTVAAVTVRALAISRG---
TH_4206|M.thermoresistible__bu FGVLSLGSALALAGVSAWLITRAWQMPPVLHLTVAVTTVRALGISRG---
MAB_2633|M.abscessus_ATCC_1997 FGVLALGSALGLAGLSAWLITRAWQMPPVLDLSVAVVAVRALGISRG---
MSMEG_5661|M.smegmatis_MC2_155 VSVLSAVLAVTTPVLAGRVIDQITDGGSAAVVLLLAGTIAAIAVAEA---
MLBr_01113|M.leprae_Br4923 LGITGTAIGIIVPWVLGHTTDLLFNGVIGRQLPKGISKTQAVAAARTRGD
.. .: . : . : : .. :.
Mb1646c|M.bovis_AF2122/97 --------------------------------------VLHYCERLATHD
Rv1620c|M.tuberculosis_H37Rv --------------------------------------VLHYCERLATHD
MAV_3167|M.avium_104 --------------------------------------VLHYCERLVTHD
MMAR_2423|M.marinum_M --------------------------------------VLHYCERLATHD
MUL_1601|M.ulcerans_Agy99 --------------------------------------VLHYCERLATHD
Mflv_3588|M.gilvum_PYR-GCK --------------------------------------LLGYCERLASHD
Mvan_2827|M.vanbaalenii_PYR-1 --------------------------------------VLGYCERLASHD
TH_4206|M.thermoresistible__bu --------------------------------------VFHYCERLASHD
MAB_2633|M.abscessus_ATCC_1997 --------------------------------------IWRYCERLAGHD
MSMEG_5661|M.smegmatis_MC2_155 --------------------------------------AVSLVTRWLSST
MLBr_01113|M.leprae_Br4923 NALADLLSGMNLVPGQGVDFVAVGRTLGLALSLYLVYTLLTWAQARLLNV
Mb1646c|M.bovis_AF2122/97 TALRAAGRARTLIYHRLAHGPAAAAVGLHSGDLAARVGADVDELANMLVR
Rv1620c|M.tuberculosis_H37Rv TALRAAGRARTLIYHRLAHGPAAAAVGLHSGDLAARVGADVDELANMLVR
MAV_3167|M.avium_104 TALRAAGAARVAVYQRLTRGGAAAAVRLHSGELVARVGADVDELANVLVR
MMAR_2423|M.marinum_M IALRAAGTARARIYQRLAEGPVAAAFRLSSGDLVARVGSDVDTLADVLVR
MUL_1601|M.ulcerans_Agy99 IALRAAVTARARIYQRLAEGPVAAAFRLSSGDLVARVGSDVDTLADVLVR
Mflv_3588|M.gilvum_PYR-GCK TALRAAGTAREQIFTRLAGGPPDAVMRRSSGDLVSRVGQGVDDLADVIVR
Mvan_2827|M.vanbaalenii_PYR-1 TALRAAATARERLFVRLATGPEDVVMRRSSGDLVSRVGAGVDELSDVVVR
TH_4206|M.thermoresistible__bu TALRAAEHARTTLYTRLAEAPEHVVSRFSSGELVARVGDSVDEVADVLVR
MAB_2633|M.abscessus_ATCC_1997 IALRGAATVRTHVYARLTGGP---AGRLRKGAVLERIGTDIDELAETVVR
MSMEG_5661|M.smegmatis_MC2_155 IGEGLILDLRTAVFDHVQKMPIAFFTRTRTGALVSRLGNDVIGAQRAFSD
MLBr_01113|M.leprae_Br4923 TVQRIMFALRSDVEDKVHRLPLSYCDTRQRGELLSRVTNDIDNLQSSLSI
* : :: * : *: .: .
Mb1646c|M.bovis_AF2122/97 ALVPIAVAAVLAVAATAVVAAVSVPAAVVLAVCLLVAGVVAPWLAGRTAA
Rv1620c|M.tuberculosis_H37Rv ALVPIAVAAVLAVAATAVVAAVSVPAAVVLAVCLLVAGVVAPWLAGRTAA
MAV_3167|M.avium_104 ALVPMGVAAVLAAAATAVVAAISPAAGGALAICLLVAGFVAPRLAGRAAA
MMAR_2423|M.marinum_M ALVPVGVAAVLAPAAIGVVAVISLPAAAVLTMCFIVAGIIAPSLARRAAQ
MUL_1601|M.ulcerans_Agy99 ALVPVDVAAVLAPAAIGVVAVISLPAAAVLTMCFIVAGIIAPSLARRAAQ
Mflv_3588|M.gilvum_PYR-GCK AVVPIGVAAVLAIAAVTTIAVISPAAAVVLALSMAVAGVVAPVLAARAVT
Mvan_2827|M.vanbaalenii_PYR-1 AVVPICVAAVLGVAAVVAIAVMSPAAAAVLAVCLLIAGVVAPALAARAAS
TH_4206|M.thermoresistible__bu AVLPIAVAVVLAVAAVSVIAVISPAAAVVLAVCLLIAGVLAPRMAARAVT
MAB_2633|M.abscessus_ATCC_1997 SVIPAAVAAVLGIAATAVVALISIPAAAVMACALLVADVLAPALAARAAR
MSMEG_5661|M.smegmatis_MC2_155 TFS--GIVTNVVTLTLTLVVMLRISWQITLLALVLVPLFVVPARRIGSRM
MLBr_01113|M.leprae_Br4923 TLS--QLLTSLLTVVAVLVVMVSVSPLLALVALLTVPMSLLATRAIAQRS
:. : . : . :. : . .: . :. : .
Mb1646c|M.bovis_AF2122/97 AQEAIARQHRGMRDTSAMIALEHAPELRVAGALRNVIADSQRRQHAWADA
Rv1620c|M.tuberculosis_H37Rv AQEAIARQHRGMRDTSAMIALEHAPELRVAGALRNVIADSQRRQHAWADA
MAV_3167|M.avium_104 TQEDIARQQHSQRDRAAMLALEHAPELRIAGLLPEVIAESQRRQRAWGAA
MMAR_2423|M.marinum_M SQEALARQHHAGRDESAMVALEHAPELRVGGVLPAVIAKSQRQQHAWGEA
MUL_1601|M.ulcerans_Agy99 SQEALARQHHAGRDESAMVALEHAPELRVGGVLPAVIAKSQRQQHAWGEA
Mflv_3588|M.gilvum_PYR-GCK ATETVAAEHHAHRDTAVMLALEHAPELRVSGRLDQVLATAGAEQRRWGHA
Mvan_2827|M.vanbaalenii_PYR-1 AAETVAVEHHAQRDNATMLALEHAPELRVSGRLDDVVAAAGDEQRRWGRA
TH_4206|M.thermoresistible__bu STEAVAAQYQSDRNTAAMLALQHAPELRVSGRLEAVITEAERRHHAWGDA
MAB_2633|M.abscessus_ATCC_1997 ARETRGARARALHSETAVEILDHAPELRVGGLLPHQLEVARHRRGEWAAA
MSMEG_5661|M.smegmatis_MC2_155 ADLSRRAAAHNATMNTQMTERFSAPGATLVKLFGSPQRESDEFAARADAV
MLBr_01113|M.leprae_Br4923 QQLFVAQWTSAGRLNAHIEETYSG--LTVVKTFGHQTAAREQFRNVNNDV
: : . : : .
Mb1646c|M.bovis_AF2122/97 LDAAARTGAIAEAMP-----TAAIGASLLGAVVAGIGMAPTVAPTTLAIL
Rv1620c|M.tuberculosis_H37Rv LDAAARTGAIAEAMP-----TAAIGASLLGAVVAGIGMAPTVAPTTLAIL
MAV_3167|M.avium_104 LDAAGKPAAVAEAMP-----TAAIGASVLGAVVAAIGMAHAVAPTTLAVL
MMAR_2423|M.marinum_M MDAAARPAAIAEAVP-----TAAIGTSVLGAVVAGILMAPTVAPTTLAVL
MUL_1601|M.ulcerans_Agy99 MDAAARPAAIAEAVP-----TAAIGTSVLGAVVAGIIMAPTVAPTTLAVL
Mflv_3588|M.gilvum_PYR-GCK LDRAAAPAAVAAAVP-----TAALGVSVVGAVLAGIALASTVAPTTVAIL
Mvan_2827|M.vanbaalenii_PYR-1 VDRAAAPAAVAAAVP-----TVAVGVSVLGAVLAGIALSHTAAPTTVAIL
TH_4206|M.thermoresistible__bu ADRAAAPAALANAAT-----TLAIGAAVLGAVVAAIPLAXXXX-------
MAB_2633|M.abscessus_ATCC_1997 QDRAAAPAAWSAAAP-----TIAVGMASVGALAAAVELANHVAPTTLAIL
MSMEG_5661|M.smegmatis_MC2_155 RDIGVRTAMLQSTFMNSLTLMSALALALVYGLGGVLAIGGQLQAGSIVSL
MLBr_01113|M.leprae_Br4923 YQASFSAQFFSGLVSPATMFISNLGYVVVAALGGLQVATGHITLGNIQAF
: . . :. : .: .
Mb1646c|M.bovis_AF2122/97 MLLPLSAFEATVALPAAAVQLTRSRIAAARLLDLTGSNRVRETESTVSAR
Rv1620c|M.tuberculosis_H37Rv MLLPLSAFEATVALPAAAVQLTRSRIAAARLLDLTGSNRVRETESTVSAR
MAV_3167|M.avium_104 MLLPLSAFEAMTPLPAAAVQLTRSRIAARRLLELTATEPPRPAG---SVP
MMAR_2423|M.marinum_M MLLPLSAFEATNALPAAAVQLTRSRIALHRLIELAGRDVNPPTDTAIATL
MUL_1601|M.ulcerans_Agy99 MLLPLSAFEATNALPAAAVQLTRSRIALHRLIELAGRDVNPPTDTAIATL
Mflv_3588|M.gilvum_PYR-GCK MLVPLAAFEGTAALPGAAVALTRGRIAAQRLLSLTDPGNDEADD------
Mvan_2827|M.vanbaalenii_PYR-1 MLLPLAAFEATGALPGAAVALTRGRIAAQRLLTLTEP--AELGD------
TH_4206|M.thermoresistible__bu -----SARPPSPLPPDSAQGWPPSR-------------------------
MAB_2633|M.abscessus_ATCC_1997 VLLPLAAFEASTALPAAAVGIARSRASARHLQELAPDGRGAEPTDVMPSA
MSMEG_5661|M.smegmatis_MC2_155 ALLLTRMYAPLTALANARVEIASALVSFERVFEVLDLEPLIAEEPDAMPV
MLBr_01113|M.leprae_Br4923 IQYVRQFNMPLSQVAGIYNTLQSGLASAERVFDLLDEPEELPDPMLALPP
. .
Mb1646c|M.bovis_AF2122/97 LPVGTGVLAADVCCGHQ---------------------EAQSIRVTIDLP
Rv1620c|M.tuberculosis_H37Rv LPVGTGVLAADVCCGHQ---------------------EAQSIRVTIDLP
MAV_3167|M.avium_104 RPTGPGRLTADIHSGHDG--------------------AARESRVTLDLR
MMAR_2423|M.marinum_M ESPTLGQLSADIYSGHS---------------------RAHATRATIELS
MUL_1601|M.ulcerans_Agy99 ESPTLGQLSADIYSGHS---------------------RAHATRTTIELS
Mflv_3588|M.gilvum_PYR-GCK ---------------------------------------RRPVVTPPVLE
Mvan_2827|M.vanbaalenii_PYR-1 ---------------------------------------RRPTVIPLHLE
TH_4206|M.thermoresistible__bu --------------------------------------------------
MAB_2633|M.abscessus_ATCC_1997 AAGAAVECSELHWHTGN----------------------SVSGPLSFRLP
MSMEG_5661|M.smegmatis_MC2_155 PDGPVTVEFDDVRFSYPSADKVSLASLEEVAELDHRGGDEVLHGISFRAE
MLBr_01113|M.leprae_Br4923 PHGQVPRINGRVEFQQVSFAYRTG--------------TPVIDDLSMVVE
Mb1646c|M.bovis_AF2122/97 PGARLAVTGASGAGKTTLLMTLAGLLPPVHG------------RVLLDGT
Rv1620c|M.tuberculosis_H37Rv PGARLAVTGASGAGKTTLLMTLAGLLPPVHG------------RVLLDGT
MAV_3167|M.avium_104 PGARLAVTGASGCGKTTLLMTLAGLLPPLQG------------QVVLDGV
MMAR_2423|M.marinum_M PGARLAVTGPSGSGKTTLLMTLAGLLSPLQG------------QVLLNGI
MUL_1601|M.ulcerans_Agy99 PGARLAVTGPSGSGKTTLLMTLAGLLSPLQG------------QVLLNGI
Mflv_3588|M.gilvum_PYR-GCK PGDRVAVVGPSGCGKTTLLMQTPG--------------------------
Mvan_2827|M.vanbaalenii_PYR-1 PGDRVAVVGPSGCGKTTLLMQTPG--------------------------
TH_4206|M.thermoresistible__bu --------GPP---------------------------------------
MAB_2633|M.abscessus_ATCC_1997 AGARMAVTGASGIGKTSLLTTLAGLTRPVSG------------TIRVRND
MSMEG_5661|M.smegmatis_MC2_155 PGQMVALVGSSGAGKSTIASLVARLYDVDVGGQERSDSGIRSGAVRLGGV
MLBr_01113|M.leprae_Br4923 PGSTVAIVGPTGAGKTTLVNLLMRFYDVDSG------------QILIDGV
*..
Mb1646c|M.bovis_AF2122/97 NLSDFDEDELRSAVSFFAEDAHIFATTVRDNLLTARGDCPDDELIEALDR
Rv1620c|M.tuberculosis_H37Rv NLSDFDEDELRSAVSFFAEDAHIFATTVRDNLLTARGDCPDDELIEALDR
MAV_3167|M.avium_104 ALSEFDECELARAIVFFAEDAHVFATTVRDNLLVARGDCPDDELTDALGA
MMAR_2423|M.marinum_M SLTRFDDAELRSTIGFFAEDAHIFATTVRDNLLVVRGDCTDDELVTTLDR
MUL_1601|M.ulcerans_Agy99 SLTRFDDAEFRSTIGFFAEDAHIFATTVRDNLLVVRGDCTDDELVTTLDK
Mflv_3588|M.gilvum_PYR-GCK --------------VFFAEDAHLFSTTVRDNLLVARGDATDDELLDALRR
Mvan_2827|M.vanbaalenii_PYR-1 --------------VFFAEDAHLFSTTVRDNLLVARGDATDDELRDALRL
TH_4206|M.thermoresistible__bu --------------------ARPWPHPCRD------------PSLHPWRR
MAB_2633|M.abscessus_ATCC_1997 ALADLPAAELPGLIRFFAEDAHLFATTVRDNLLVARGDAPDGDLVDALSV
MSMEG_5661|M.smegmatis_MC2_155 DVRDVTFASLKDTVGMVTQDGHLFHESIGANLRLAAPAATDDELWDALRR
MLBr_01113|M.leprae_Br4923 DITTVSRHALRSRIGMVLQDAWLFDGTIAENIAYGRPQASEDEVMEAAKA
. : . .
Mb1646c|M.bovis_AF2122/97 VGLCGWLAGLPEGLSTVLIGGAQAVSAGQRRRLLLARAVLSPARIVLLDE
Rv1620c|M.tuberculosis_H37Rv VGLCGWLAGLPEGLSTVLIGGAQAVSAGQRRRLLLARAVLSPARIVLLDE
MAV_3167|M.avium_104 VGLDDWLAGLPDGLSTVLTGGAQAISAGQRRRLLLARAVISPARIVLLDE
MMAR_2423|M.marinum_M VGLGAWLAGLPHGLSTVLIGGAQAVSAGQRRRLLLARAALSPAQIVLLDE
MUL_1601|M.ulcerans_Agy99 VGLGAWLAGLPHGLSTVLIVGAQAVSAGQRRRLLLARAALSPAQIVLLDE
Mflv_3588|M.gilvum_PYR-GCK CGLGRWLAGLSDGLSTVLVGGAAAVSAGQRRRLLLARALVSNAQTVLLDE
Mvan_2827|M.vanbaalenii_PYR-1 AGLGAWLDGLPQGLSTLLVGGAAAVSAGQRRRLLLARALVSAAPTVLLDE
TH_4206|M.thermoresistible__bu SSCPGWVS---------------------RRRFRATYARPGPP-------
MAB_2633|M.abscessus_ATCC_1997 VGLGPWLESLPQGLDTVMTAGAGSVSGGQRRRILLARALLSRVPVLLLDE
MSMEG_5661|M.smegmatis_MC2_155 ARLADVVAAMPDGLDTIVGERGYRLSGGQRQRLTIARLLLGRSRVVVLDE
MLBr_01113|M.leprae_Br4923 AYVDRFVHTLSAGYQTRVSGDGDNISAGEKQLITIARAFLARPQLLILDE
: :: : : .
Mb1646c|M.bovis_AF2122/97 PVEHLDAANAD-LLRDLLAPNS---GIMSAMRTVVVATHHLPNDIQCAEL
Rv1620c|M.tuberculosis_H37Rv PVEHLDAANAD-LLRDLLAPNS---GIMSAMRTVVVATHHLPNDIQCAEL
MAV_3167|M.avium_104 PTEHLDAGDAERLLRDLLAPDS---RLLSPPRTVVVATHHLPEGIGCREL
MMAR_2423|M.marinum_M PTEHLDATDADRILRDLLSSET---GLLPVAKTIVVATHHLSENIRCDRM
MUL_1601|M.ulcerans_Agy99 PTEHLDAIDADRILRDLLSSET---GLLPVAKTIVVATHHLSENIRCDRM
Mflv_3588|M.gilvum_PYR-GCK PTEHLDASDADAILTGLLTPG----ALFGPERTVVVATHHLPEHLDVKVI
Mvan_2827|M.vanbaalenii_PYR-1 PTEHLDASDADQILTDLLTPG----TLFSPQQTVVVATHHLPEGVPVTVV
TH_4206|M.thermoresistible__bu -----------------------------PWRTPI---------------
MAB_2633|M.abscessus_ATCC_1997 PTEHLDGRDSDSIMRKLLNAN----SSLLAGRTVVVATHHLPDSVSCLRI
MSMEG_5661|M.smegmatis_MC2_155 ATASLDSASEAAVQQALAEALEGRTSLVIAHRLSTVRAADLILVVEDGRI
MLBr_01113|M.leprae_Br4923 ATSSIDTRTEALIARATHDLLQDRTSFIIAHHLPTIRDADHILVVAAGKI
:
Mb1646c|M.bovis_AF2122/97 SIATDQRCRRRGTNSSDNNTNASAKT-----
Rv1620c|M.tuberculosis_H37Rv SIATDQRCRRRGTNSSDNNTNASAKT-----
MAV_3167|M.avium_104 RVD-------RGPAAG---------------
MMAR_2423|M.marinum_M NIPSRQEVSPKVTPARHELQ-----------
MUL_1601|M.ulcerans_Agy99 NIPSRQEVSPKVTPARHELQ-----------
Mflv_3588|M.gilvum_PYR-GCK SPT----------------------------
Mvan_2827|M.vanbaalenii_PYR-1 SPT----------------------------
TH_4206|M.thermoresistible__bu -------------------------------
MAB_2633|M.abscessus_ATCC_1997 QLGDTVQV-----------------------
MSMEG_5661|M.smegmatis_MC2_155 VERGTHEALLARAGRYAELYYTQFDTDASAA
MLBr_01113|M.leprae_Br4923 VEQGSHAKLLARRGAYYRMTQA---------