For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGQATKLAGAVDSRYRASAGLRRQLNKVFPTHWSFLLGEIAVYSFVVLLLSGIYLALFFDPSMSEIAYNG VYQPLRGTEMSTAYASALDLSFEVRGGLFVRQIHHWAALMFAASIVVHLARVFFTGAFRRPREANWVIGS VLLILAMAEGFTGYSLPDDLLSGTGLRVVTQGMMSLPIVGTWMHWGLFGGDFPGHSVLPRLYVLHVLLVP AIMIALISAHLVLVWYQKHTQFPGPRRTESNVVGVRIVPVFALKSGALFAVVVAILALMGGLLQINPVWN LGPYKPSQVAAGSPPDIYLMWTDGLLRLVPNWELYVFGHTVPAVLWGVLGILAAFMVIVAWPWLEQHFTG DDAAHNLLQRPRDAPVRTAIGAAAFSLYFLLTLSCMNDIIALKFHISLNATTWIGRIGMVILPAVVYYIA YRWAIGLQRSDRAVLEHGIETGIIKRLPHGEYIEIHQPLAGVDEHGHAIPLEYQGAPVPQRMNKLGSAGA PGTGSFLFADPADEQHALAEAEHEAMCAQLAALRQ
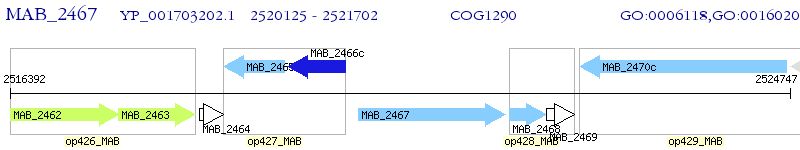
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2467 | - | - | 100% (525) | ubiquinol-cytochrome c reductase cytochrome b subunit |
| M. abscessus ATCC 19977 | MAB_1966c | - | 0.0 | 78.05% (524) | ubiquinol-cytochrome c reductase cytochrome b subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2219 | qcrB | 0.0 | 73.06% (516) | ubiquinol-cytochrome C reductase QcrB (cytochrome B subunit) |
| M. gilvum PYR-GCK | Mflv_2953 | - | 0.0 | 72.47% (523) | cytochrome b/b6 domain-containing protein |
| M. tuberculosis H37Rv | Rv2196 | qcrB | 0.0 | 73.06% (516) | ubiquinol-cytochrome C reductase QcrB (cytochrome B subunit) |
| M. leprae Br4923 | MLBr_00879 | qcrB | 0.0 | 71.51% (516) | putative cytochrome B |
| M. marinum M | MMAR_3240 | qcrB | 0.0 | 70.53% (526) | ubiquinol-cytochrome C reductase QcrB |
| M. avium 104 | MAV_2296 | - | 0.0 | 69.96% (526) | cytochrome b6 |
| M. smegmatis MC2 155 | MSMEG_4263 | - | 0.0 | 73.11% (517) | ubiquinol-cytochrome c reductase cytochrome b subunit |
| M. thermoresistible (build 8) | TH_1044 | qcrB | 0.0 | 69.60% (523) | Probable Ubiquinol-cytochrome C reductase QcrB (cytochrome B |
| M. ulcerans Agy99 | MUL_3553 | qcrB | 0.0 | 70.53% (526) | ubiquinol-cytochrome C reductase QcrB |
| M. vanbaalenii PYR-1 | Mvan_3560 | - | 0.0 | 72.28% (523) | cytochrome b/b6 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3240|M.marinum_M MSPKLSPPKIGDVLARQAEDIDTRYHPAAALRRQFNKVFPTHWSFLLGEV
MUL_3553|M.ulcerans_Agy99 MSPKLSPPKIGDVLARQAEDIDTRYHPAAALRRQFNKVFPTHWSFLLGEV
MAV_2296|M.avium_104 MSPKLSPPKIGDVLARQGEDIDTRYHPSAAVRRQLNKVFPTHWSFLLGEI
Mb2219|M.bovis_AF2122/97 MSPKLSPPNIGEVLARQAEDIDTRYHPSAALRRQLNKVFPTHWSFLLGEI
Rv2196|M.tuberculosis_H37Rv MSPKLSPPNIGEVLARQAEDIDTRYHPSAALRRQLNKVFPTHWSFLLGEI
MLBr_00879|M.leprae_Br4923 MSPK-SVPDIGDVLARQAEDIDTRYHPSAALRRQLNKVFPTHWSFLLGEI
Mflv_2953|M.gilvum_PYR-GCK MSPR---LDAATVAAKQGDAIDSRYHPSAAVRRQLNKVFPTHWSFLLGEI
Mvan_3560|M.vanbaalenii_PYR-1 MSPR---PTAAEIAARQGNAIDSRYHPSAAVRRQLNKVFPTHWSFLLGEI
MSMEG_4263|M.smegmatis_MC2_155 MSP-----DFAKLAAAQGDAIDSRYHPSAAVRRQLNKVFPTHWSFLLGEI
TH_1044|M.thermoresistible__bu MSP-----KLAEIAAKQGEAIDSRYHPSAAVRRQLNKVFPTHWSFLLGEI
MAB_2467|M.abscessus_ATCC_1997 ----------MGQATKLAGAVDSRYRASAGLRRQLNKVFPTHWSFLLGEI
: . :*:**:.:*.:***:**************:
MMAR_3240|M.marinum_M ALYSFIVLLITGVYLTLFFDPSMMDVTYNGVYQPLRGVEMSKAYASALDI
MUL_3553|M.ulcerans_Agy99 ALYSFIVLLITGVYLTLFFDPSMMDVTYNGVYQPLRGVEMSKAYASALDI
MAV_2296|M.avium_104 AMYSFIVLLLTGVYLTLFFDPSMAEVTYNGVYQPLRGVEMSKAFASALDI
Mb2219|M.bovis_AF2122/97 ALYSFVVLLITGVYLTLFFDPSMVDVTYNGVYQPLRGVEMSRAYQSALDI
Rv2196|M.tuberculosis_H37Rv ALYSFVVLLITGVYLTLFFDPSMVDVTYNGVYQPLRGVEMSRAYQSALDI
MLBr_00879|M.leprae_Br4923 ALYSFIVLLLTGVYLTLFFDPSMTDVTYNGVYQPLRGVEMSRAYQSTLDI
Mflv_2953|M.gilvum_PYR-GCK AMYSFIVLLLTGVYLTLFFDPSMAHVTYDGVYQPLRGVGMSRAYETTLDI
Mvan_3560|M.vanbaalenii_PYR-1 AMYSFIVLLLTGVWLTLFFDPSMAHVTYDGVYQPLRGVGMSRAYETTLDI
MSMEG_4263|M.smegmatis_MC2_155 ALYSFIILLLTGVWLTLFFDPSMAHVTYDGVYQPLRGVQMSRAYETALDI
TH_1044|M.thermoresistible__bu ALYSFIVLIITGIYLTLFFDPSMAHVVYDGVYQPLRGVEMSRAYETALNI
MAB_2467|M.abscessus_ATCC_1997 AVYSFVVLLLSGIYLALFFDPSMSEIAYNGVYQPLRGTEMSTAYASALDL
*:***::*:::*::*:******* .:.*:********. ** *: ::*::
MMAR_3240|M.marinum_M SFEVRGGLFVRQVHHWAALMFAAAIMVHLARIFFTGAFRRPREANWIIGS
MUL_3553|M.ulcerans_Agy99 SFEVRGGLFVRQVHHWAALMFAAAIMVHLARIFFTGAFRRPREANWIIGS
MAV_2296|M.avium_104 SFEVRGGLFVRQVHHWAALIFSAAIMVHLARIFFTGAFRRPREANWVIGS
Mb2219|M.bovis_AF2122/97 SFEVRGGLFVRQIHHWAALMFAAAIMVHLARIFFTGAFRRPRETNWVIGS
Rv2196|M.tuberculosis_H37Rv SFEVRGGLFVRQIHHWAALMFAAAIMVHLARIFFTGAFRRPRETNWVIGS
MLBr_00879|M.leprae_Br4923 SFEVRGGLFVRQIHHWAALMFTAAIMVHLARIFFTGAFRRPRETNWVIGS
Mflv_2953|M.gilvum_PYR-GCK SFEVRGGLFVRQVHHWAALLFAASIMVHLARIFFTGAFRRPREANWVIGS
Mvan_3560|M.vanbaalenii_PYR-1 SFEVRGGLFVRQVHHWAALLFAASIMVHLARIFFTGAFRRPREANWVIGS
MSMEG_4263|M.smegmatis_MC2_155 SFEVRGGLFVRQVHHWAALMFAASIMVHLARIFFTGAFRRPREANWVIGS
TH_1044|M.thermoresistible__bu SFEVRGGLFVRQIHHWAALLFAAAIMVHLARVFFTGAFRRPREANWVIGC
MAB_2467|M.abscessus_ATCC_1997 SFEVRGGLFVRQIHHWAALMFAASIVVHLARVFFTGAFRRPREANWVIGS
************:******:*:*:*:*****:***********:**:**.
MMAR_3240|M.marinum_M LLLILAMFEGYFGYSLPDDLLSGIGLRAALSSITLGMPVIGTWLHWALFG
MUL_3553|M.ulcerans_Agy99 LLLILAMFEGYFGYSLPDDLLSGIGLRAALSSITLGMPVIGTWLHWALFG
MAV_2296|M.avium_104 LLLILAMFEGYFGYSLPDDLLSGIGLRAALSSITLGMPVIGTWLHWALFG
Mb2219|M.bovis_AF2122/97 LLLILAMFEGYFGYSLPDDLLSGLGLRAALSSITLGMPVIGTWLHWALFG
Rv2196|M.tuberculosis_H37Rv LLLILAMFEGYFGYSLPDDLLSGLGLRAALSSITLGMPVIGTWLHWALFG
MLBr_00879|M.leprae_Br4923 LLLILAMFEGYFGYSMPDDLLSGIGLRAALSSITLGIPVIGTWLHWALFG
Mflv_2953|M.gilvum_PYR-GCK LLLILAMFEGFFGYSLPDDLLSGTGIRAALSGITMGIPVVGTWMHWALFG
Mvan_3560|M.vanbaalenii_PYR-1 LLLILAMFEGFFGYSLPDDLLSGTGIRAALSGITMGIPIVGTWMHWALFG
MSMEG_4263|M.smegmatis_MC2_155 LLLILAMFEGFFGYSLPDDLLSGTGIRAALSGITMGIPVIGTWMHWALFG
TH_1044|M.thermoresistible__bu LLLILAMFEGFFGYTLPDELLSGTGLRASFSSITLGMPVIGTWLHWMLFG
MAB_2467|M.abscessus_ATCC_1997 VLLILAMAEGFTGYSLPDDLLSGTGLRVVTQGMMS-LPIVGTWMHWGLFG
:****** **: **::**:**** *:*. ..: :*::***:** ***
MMAR_3240|M.marinum_M GDFPCGGVGDDCTAAGYIIPRMYSLHILLLPGIILALIGMHMALVWFQKH
MUL_3553|M.ulcerans_Agy99 GDFPCGGVGDDCTAAGYIIPRMYSLHILLLPGIILALIGMHMALVWFQKH
MAV_2296|M.avium_104 GDFPCGGVGSECAAAGYIIPRMYALHILLLPGIILALIGLHLAMVWFQKH
Mb2219|M.bovis_AF2122/97 GDFPG----------TILIPRLYALHILLLPGIILALIGLHLALVWFQKH
Rv2196|M.tuberculosis_H37Rv GDFPG----------TILIPRLYALHILLLPGIILALIGLHLALVWFQKH
MLBr_00879|M.leprae_Br4923 GDFPG----------TILIPRLYALHILLIPGVILALIGLHLALVWFQKH
Mflv_2953|M.gilvum_PYR-GCK GDFPG----------EILIPRLYALHILLIPGIILALIGVHLALVWFQKH
Mvan_3560|M.vanbaalenii_PYR-1 GDFPG----------DILIPRLYALHILLIPGIILALIGVHLALVWFQKH
MSMEG_4263|M.smegmatis_MC2_155 GDFPG----------EILIPRLYALHILLIPGIILALIGAHLALVWFQKH
TH_1044|M.thermoresistible__bu GDFPG----------NIIIPRMYALHILILPGIMLALVGAHLALVWFQKH
MAB_2467|M.abscessus_ATCC_1997 GDFPG----------HSVLPRLYVLHVLLVPAIMIALISAHLVLVWYQKH
**** ::**:* **:*::*.:::**:. *:.:**:***
MMAR_3240|M.marinum_M TQFPGPGRTEHNVVGVRVMPVFAVKSGAFFAAITGVLGLMGGLLQINPIW
MUL_3553|M.ulcerans_Agy99 TQFPGPGRTEHNVVGVRVMPVFAVKSGAFFAAITGVLGLMGGLLQINPIW
MAV_2296|M.avium_104 TQFPGPGRTEHNVVGVRVMPVFAVKSGAFFAAIVGVLGLMGGLLQINPIW
Mb2219|M.bovis_AF2122/97 TQFPGPGRTEHNVVGVRVMPVFAFKSGAFFAAIVGVLGLMGGLLQINPIW
Rv2196|M.tuberculosis_H37Rv TQFPGPGRTEHNVVGVRVMPVFAFKSGAFFAAIVGVLGLMGGLLQINPIW
MLBr_00879|M.leprae_Br4923 TQFPGPGRTEYNVVGVRVMPVFAFKSGAFFAAIVGVLGLMGGFLQINPIW
Mflv_2953|M.gilvum_PYR-GCK TQFPGPGRTETNVVGVRVMPVFAVKSGAFFAVTVGILGLMGGLLQINPVW
Mvan_3560|M.vanbaalenii_PYR-1 TQFPGPGRTETNVVGVRVMPVFAVKSGAFFAMTVGVLGLMGGLLQINPVW
MSMEG_4263|M.smegmatis_MC2_155 TQFPGPGRTETNVVGVRVMPVFAVKSGAFFAMITGVLGLMGGLLTINPIW
TH_1044|M.thermoresistible__bu TQFPGPGRTETNVVGVRVMPVFAVKSGAFFAMTVGVLGIMGGLLQINPIW
MAB_2467|M.abscessus_ATCC_1997 TQFPGPRRTESNVVGVRIVPVFALKSGALFAVVVAILALMGGLLQINPVW
****** *** ******::****.****:** ..:*.:***:* ***:*
MMAR_3240|M.marinum_M NLGPYKPAHVSAGSQPDFYMMWTEGLARIWPPWEFYFWHHTIPAPVWVAL
MUL_3553|M.ulcerans_Agy99 NLGPYKPAHVSAGSQPDFYMMWTEGLARIWPPWEFYFWHHTIPAPVWVAL
MAV_2296|M.avium_104 NLGPYKPSHVSAGSQPDFYMMWTEGLARIWPPWEFYFWHHTIPAVVWVAL
Mb2219|M.bovis_AF2122/97 NLGPYKPSQVSAGSQPDFYMMWTEGLARIWPPWEFYFWHHTIPAPVWVAV
Rv2196|M.tuberculosis_H37Rv NLGPYKPSQVSAGSQPDFYMMWTEGLARIWPPWEFYFWHHTIPAPVWVAV
MLBr_00879|M.leprae_Br4923 NLGPYKPSQVSAGSQPDFYMMWTEGLARIWPAWEFYFWHHTIPAPVWVAV
Mflv_2953|M.gilvum_PYR-GCK NIGPYNPSQVSAGSQPDFYMMWTDGLIRLWPAWEFYPFGHTIPQGVWVAL
Mvan_3560|M.vanbaalenii_PYR-1 TIGPYKPSQVSAGSQPDFYMMWTDGLIRLWPAWEFYPFGHTIPQGVWVAV
MSMEG_4263|M.smegmatis_MC2_155 NLGPYKPSQVSAGSQPDFYMMWTDGLIRLWPAWEFYPFGHTIPQGVWVAV
TH_1044|M.thermoresistible__bu NLGPYRPSQISAGSQPDFYMMWTEGMARIFPPWEIYIGNYTIPASTWVAL
MAB_2467|M.abscessus_ATCC_1997 NLGPYKPSQVAAGSPPDIYLMWTDGLLRLVPNWELYVFGHTVPAVLWGVL
.:***.*::::*** **:*:***:*: *: * **:* :*:* * .:
MMAR_3240|M.marinum_M IMGLIFMLLIVYPFLEKRFTGDYAHHNLLQRPRDAPVRTAVGAMAISFYM
MUL_3553|M.ulcerans_Agy99 IMGLIFMLLIVYPFLEKRFTGDYAHHNLLQRPRDAPVRTAVGAMAISFYM
MAV_2296|M.avium_104 IMGAVFVLLIIYPFLEKRFSGDYAHHNLLQRPRDVPVRTSIGAMAIAFYM
Mb2219|M.bovis_AF2122/97 IMGLVFVLLPAYPFLEKRFTGDYAHHNLLQRPRDVPVRTAIGAMAIAFYM
Rv2196|M.tuberculosis_H37Rv IMGLVFVLLPAYPFLEKRFTGDYAHHNLLQRPRDVPVRTAIGAMAIAFYM
MLBr_00879|M.leprae_Br4923 IMALVFVLLITYPFLEKRFTGDYAHHNLLQRPRDVPVRTSIGAMAITFYM
Mflv_2953|M.gilvum_PYR-GCK AMGLIFMLLGAYPFIEKKLTGDNAHHNLLQRPRDAPTRTAVGAAAIAFYI
Mvan_3560|M.vanbaalenii_PYR-1 GMGLIFTLLAAYPFIEKKLSGDDAHHNLLQRPRDAPTRTAVGAAAIAFYI
MSMEG_4263|M.smegmatis_MC2_155 GMGLVFALLIAYPFIEKKVTGDDAHHNLLQRPRDVPVRTAIGSMAIALYL
TH_1044|M.thermoresistible__bu AMAVVFIGLIVYPWVEKKLTGDDAHHNLLQRPRDVPVRTGIGAAAISFYL
MAB_2467|M.abscessus_ATCC_1997 GILAAFMVIVAWPWLEQHFTGDDAAHNLLQRPRDAPVRTAIGAAAFSLYF
: * : :*::*::.:** * *********.*.**.:*: *:::*:
MMAR_3240|M.marinum_M LLTLAAMNDIIALKFHISLNATTWIGRIGMVILPPFVYFISYRWSIGLQR
MUL_3553|M.ulcerans_Agy99 LLTLAAMNDIIALKFHISLNATTWIGRIGMVILPPFVYFISYRWSIGLQR
MAV_2296|M.avium_104 VLTLSAMNDIIALKFDISLNATTWIGRIGMVIVPPIVYFVTYRWCIGLQR
Mb2219|M.bovis_AF2122/97 VLTLAAMNDIIALKFHISLNATTWIGRIGMVILPPFVYFITYRWCIGLQR
Rv2196|M.tuberculosis_H37Rv VLTLAAMNDIIALKFHISLNATTWIGRIGMVILPPFVYFITYRWCIGLQR
MLBr_00879|M.leprae_Br4923 VLTLAAMNDIIALKFHISLNATTWIGRIGMVILPLLVYFITYRWCIGLQR
Mflv_2953|M.gilvum_PYR-GCK VLTFACMNDIIALKFHISLNATTWIGRVGMVLLPAIVYFVTYRWAVALQR
Mvan_3560|M.vanbaalenii_PYR-1 VLTFACMNDIIALKFHISLNATTWIGRVGMVVLPAIVYFVAYRWAVGLQR
MSMEG_4263|M.smegmatis_MC2_155 LLTFACMNDIIALKFHISLNATTWIGRIGMVVLPAIVYFVAYRWAISLQR
TH_1044|M.thermoresistible__bu LMTLAAMNDVIAWKFHISLNATTWIGRIGMVVLPAIVYFVAYRWAVSLQR
MAB_2467|M.abscessus_ATCC_1997 LLTLSCMNDIIALKFHISLNATTWIGRIGMVILPAVVYYIAYRWAIGLQR
::*::.***:** **.***********:***::* .**:::***.:.***
MMAR_3240|M.marinum_M SDRAVLEHGIETGIIKRLPHGAYIELHQPLGPVDEHGHPLPLDYQGAPLP
MUL_3553|M.ulcerans_Agy99 SDRAVLEHGIETGIIKRLPHGAYIELHQPLGPVDEHGHPLPLDYQGAPLP
MAV_2296|M.avium_104 SDRAVLEHGIETGIIKRLPHGAYIELHQPLGPVDEHGHPLPLEYQGAPLP
Mb2219|M.bovis_AF2122/97 SDRSVLEHGVETGIIKRLPHGAYIELHQPLGPVDEHGHPIPLQYQGAPLP
Rv2196|M.tuberculosis_H37Rv SDRSVLEHGVETGIIKRLPHGAYIELHQPLGPVDEHGHPIPLQYQGAPLP
MLBr_00879|M.leprae_Br4923 SDRAVLEHGIETGIIKRLPHGAYIELHQPLGPVDDHGHPIPLEYQGTAVP
Mflv_2953|M.gilvum_PYR-GCK SDRAVLEHGVETGIIKRLPHGAYVELHQPLGPVDDHGHPLPLEYQGAALP
Mvan_3560|M.vanbaalenii_PYR-1 SDRAVLEHGVETGIIKRLPHGAYVELHQPLGPVDEHGHPIPLEYQGAALP
MSMEG_4263|M.smegmatis_MC2_155 SDREVLEHGVETGIIKRLPHGAYVELHQPLGPVDEHGHPIPLEYAGAPLP
TH_1044|M.thermoresistible__bu SDREVLEHGVETGIIKRLPHGAYIELHQPLGPVDEHGHPIPLEYQGAPVP
MAB_2467|M.abscessus_ATCC_1997 SDRAVLEHGIETGIIKRLPHGEYIEIHQPLAGVDEHGHAIPLEYQGAPVP
*** *****:*********** *:*:****. **:***.:**:* *:.:*
MMAR_3240|M.marinum_M KRMNKLGSAGSPGSGSFLTADPASEDAALREAGHAAEHRALTALREYQDS
MUL_3553|M.ulcerans_Agy99 KRMNKLGSAGSPGSGSFLTADPASEDAALREAGHAAEHRALTALREYQDS
MAV_2296|M.avium_104 KKMNKLGSAGSPGIGSFLYADPPSEDAALREAAHASEQRALTALREHQDS
Mb2219|M.bovis_AF2122/97 KRMNKLGSAGSPGSGSFLFADSAAEDAALREAGHAAEQRALAALREHQDS
Rv2196|M.tuberculosis_H37Rv KRMNKLGSAGSPGSGSFLFADSAAEDAALREAGHAAEQRALAALREHQDS
MLBr_00879|M.leprae_Br4923 KRMNKLGSAGSPSSGSFLFADPVSEDAALREATHVAEQRALTALREHQDS
Mflv_2953|M.gilvum_PYR-GCK KRMNKLGSAGAPGSGSFLKADPLSEHEAIAEATHAAEHRALTALREHQER
Mvan_3560|M.vanbaalenii_PYR-1 KRMNKLGSAGAPGSGSFLKGDPLSEHEALDEAAHAAERRALTALREHQER
MSMEG_4263|M.smegmatis_MC2_155 KRMNKLGSGGAPGTGSFLFPDPAVEHEALTEAAHASEHKSLTALKEHQDR
TH_1044|M.thermoresistible__bu KRMNKLGAGGVPGTGSFITPDPISEHEALVEAAHASERKALAALEEYQEN
MAB_2467|M.abscessus_ATCC_1997 QRMNKLGSAGAPGTGSFLFADPADEQHALAEAEHEAMCAQLAALRQ----
::*****:.* *. ***: *. *. *: ** * : *:**.:
MMAR_3240|M.marinum_M LNE-------TSNG-----EGDH
MUL_3553|M.ulcerans_Agy99 LNE-------TSNG-----EGDH
MAV_2296|M.avium_104 IVG-------STNGSTNGEDGQH
Mb2219|M.bovis_AF2122/97 IMG-------SPDG----EH---
Rv2196|M.tuberculosis_H37Rv IMG-------SPDG----EH---
MLBr_00879|M.leprae_Br4923 IAS-------SPNG----ERGKH
Mflv_2953|M.gilvum_PYR-GCK SSAPNGSSDGASNGSS---NGHH
Mvan_3560|M.vanbaalenii_PYR-1 TSA--------SNGSS---NGHH
MSMEG_4263|M.smegmatis_MC2_155 IHG---------NGET---NGHH
TH_1044|M.thermoresistible__bu LRD------GQGNGEV---ARR-
MAB_2467|M.abscessus_ATCC_1997 -----------------------