For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLVLVMLPLLPLIRTETMSEEGIRLQKVLSQAGIASRRVAERMIYDGRVEVDGEIVTEQGRRIDPAVNVV RVDGARVIMNTDLVYLALNKPFGMLSTMSDDRGRPCVGKLIEERVRGNQGVRNVGRLDADTVGLILLTND GELTHRLMHPSYEVPKTYIATVAGTIPRGLGRQLRDGVELADGPAKVDEFKFLGTTDGQSMVQLTLHEGR NRIVRRMMDAAGHPVVELVRTKIGDVTLGNQRPGSLRALGRDEIGSLYKAVGL
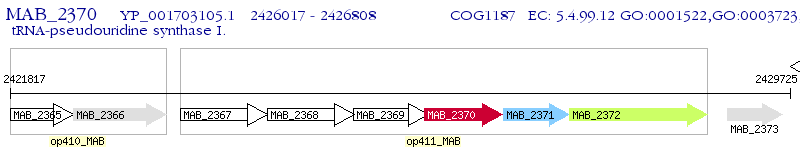
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2370 | - | - | 100% (263) | l RNA pseudouridine synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1738 | - | 2e-96 | 70.16% (248) | hypothetical protein Mb1738 |
| M. gilvum PYR-GCK | Mflv_3495 | - | 4e-96 | 69.96% (243) | pseudouridine synthase |
| M. tuberculosis H37Rv | Rv1711 | - | 2e-96 | 70.16% (248) | hypothetical protein Rv1711 |
| M. leprae Br4923 | MLBr_01370 | - | 6e-92 | 68.88% (241) | putative pseudouridine synthase |
| M. marinum M | MMAR_2526 | - | 5e-98 | 71.31% (244) | pseudouridylate synthase |
| M. avium 104 | MAV_3065 | - | 4e-97 | 70.37% (243) | ribosomal large subunit pseudouridine synthase B |
| M. smegmatis MC2 155 | MSMEG_3740 | rluB | 2e-99 | 71.72% (244) | ribosomal large subunit pseudouridine synthase B |
| M. thermoresistible (build 8) | TH_0200 | - | 1e-96 | 68.44% (244) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3234 | - | 3e-98 | 71.31% (244) | pseudouridylate synthase |
| M. vanbaalenii PYR-1 | Mvan_3276 | - | 4e-97 | 70.37% (243) | pseudouridine synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2526|M.marinum_M -----MTDTEDFEQER-----GIRLQKVLSQAGIASRRAAEKLIIDGRVE
MUL_3234|M.ulcerans_Agy99 -----MTDTEDFEQER-----GIRLQKVLSQAGIASRRAAEKLIIDGRVE
Mb1738|M.bovis_AF2122/97 ----MMAEPEESREPR-----GIRLQKVLSQAGIASRRAAEKMIVDGRVE
Rv1711|M.tuberculosis_H37Rv ----MMAEPEESREPR-----GIRLQKVLSQAGIASRRAAEKMIVDGRVE
MAV_3065|M.avium_104 ----MMVEAQ-----------GIRLQKVLSQAGIASRRAAEKLIVDGRVE
MLBr_01370|M.leprae_Br4923 ----MMVEIHDSRMDRGAG--AVRLQKILSRAGIASRRAAEKLIIEGRVE
Mflv_3495|M.gilvum_PYR-GCK -----MTDND-----------GVRLQKVLSQAGIASRRVAEKMIRDGRVE
Mvan_3276|M.vanbaalenii_PYR-1 -----MTDND-----------GIRLQKVLSQAGIASRRVAEKMIRDGRVE
TH_0200|M.thermoresistible__bu -----MADQD-----------GVRLQKVLSQAGVASRRVAEQMIREGRVE
MSMEG_3740|M.smegmatis_MC2_155 -----MAEEQ-----------GVRLQKVLSQAGIASRRVAERMIIDGRVE
MAB_2370|M.abscessus_ATCC_1997 MLVLVMLPLLPLIRTETMSEEGIRLQKVLSQAGIASRRVAERMIYDGRVE
* .:****:**:**:****.**::* :****
MMAR_2526|M.marinum_M VDGQVVTELGTRVDPDTSLIRVDGVRVILDDSLVYLALNKPRGMHSTMSD
MUL_3234|M.ulcerans_Agy99 VDGQVVTELGTRVDPDTSLIRVDGVRVILDDSLVYLALNKPRGMHSTMSD
Mb1738|M.bovis_AF2122/97 VDGHVVTELGTRVDPQVAVVRVDGARVVLDDSLVYLALNKPRGMHSTMSD
Rv1711|M.tuberculosis_H37Rv VDGHVVTELGTRVDPQVAVVRVDGARVVLDDSLVYLALNKPRGMHSTMSD
MAV_3065|M.avium_104 VDGQVVTELGTRVDPDAAVIRVDGARVVLDESQVYLALNKPRGVHSTMSD
MLBr_01370|M.leprae_Br4923 VDGQLVRELGTRVDPDVSVVRVDGVKVVVDDSLVYLALNKPRGMYSTMSD
Mflv_3495|M.gilvum_PYR-GCK VDDRIVTELGTRVDPAESVIRVDGARITVDDTLVHLAINKPRGMHSTMSD
Mvan_3276|M.vanbaalenii_PYR-1 VDGRIVTELGTRVDPAASVIRVDGARITVDDTLVYLAINKPRGMHSTMSD
TH_0200|M.thermoresistible__bu VDGRIVTELGTRVDPDTAEIRVDGMRIVLDDTMVYLAINKPRGMHSTMSD
MSMEG_3740|M.smegmatis_MC2_155 VDGQIVTELGTRVDPAVADIRVDGSRIILDDTMVYLAINKPRGMHSTMSD
MAB_2370|M.abscessus_ATCC_1997 VDGEIVTEQGRRIDPAVNVVRVDGARVIMNTDLVYLALNKPFGMLSTMSD
**..:* * * *:** :**** :: :: *:**:*** *: *****
MMAR_2526|M.marinum_M DRGRPCIGDLIERKVRGNKNLFHVGRLDADTEGLILLTNDGELAHRLMHP
MUL_3234|M.ulcerans_Agy99 DRGRPCIGDLIERKVRGNKNLFHVGRLDADTEGLILLTNDGELAHRLMHP
Mb1738|M.bovis_AF2122/97 DRGRPCIGDLIERKVRGTKKLFHVGRLDADTEGLMLLTNDGELAHRLMHP
Rv1711|M.tuberculosis_H37Rv DRGRPCIGDLIERKVRGTKKLFHVGRLDADTEGLMLLTNDGELAHRLMHP
MAV_3065|M.avium_104 DRGRPCIGDLIERRVRGNKNLFHVGRLDAETEGLILLTNDGELAHRLMHP
MLBr_01370|M.leprae_Br4923 DRGRPCVGDLIERRVRGNKKLFHVGRLDADTEGLILLTNDGELAHRLMHP
Mflv_3495|M.gilvum_PYR-GCK DRGRPCVGDLVEHRVRGNKKLFHVGRLDADTEGLLLLTNDGELAHRLMHP
Mvan_3276|M.vanbaalenii_PYR-1 DRGRPCVGDLVEHRVRGNKKLFHVGRLDADTEGLLLLTNDGELAHRLMHP
TH_0200|M.thermoresistible__bu DRGRPCIGDLVEHRVRGNKNLFHVGRLDADTEGLMLLTNDGELAHRLMHP
MSMEG_3740|M.smegmatis_MC2_155 DRGRPCVGDLVEHRVRGNKKLFHVGRLDADTEGLLLLTNDGELAHRLMHP
MAB_2370|M.abscessus_ATCC_1997 DRGRPCVGKLIEERVRGNQGVRNVGRLDADTVGLILLTNDGELTHRLMHP
******:*.*:*.:***.: : :******:* **:********:******
MMAR_2526|M.marinum_M SHEVPKTYLATVAGTVPRGLGKKLRAGIELEDGPVRVDGFAVVDAIPGKT
MUL_3234|M.ulcerans_Agy99 SHEVPKTYLATVAGTVPRGLGKKLRAGIELEDGPVRVDGFAVVDAIPGKT
Mb1738|M.bovis_AF2122/97 SHEVPKTYLATVTGSVPRGLGRTLRAGIELDDGPAFVDDFAVVDAIPGKT
Rv1711|M.tuberculosis_H37Rv SHEVPKTYLATVTGSVPRGLGRTLRAGIELDDGPAFVDDFAVVDAIPGKT
MAV_3065|M.avium_104 SHEVPKTYLATVTGSIPRGLGKTLRAGIELDDGMARVDEFAVVDAIPGKT
MLBr_01370|M.leprae_Br4923 SHEVSKTYLATVKGAVPRGLGKKLSVGLELDDGPAHVDDFAVVDAIPGKT
Mflv_3495|M.gilvum_PYR-GCK SYEVPKSYVATVLGSVPRGLGRKLREGVELDDGPVQVDDFALVDTTPGRT
Mvan_3276|M.vanbaalenii_PYR-1 SFEVPKSYVATVLGSVPKGLGRKLREGVELEDGPVQVDDFAVVDTTPGRT
TH_0200|M.thermoresistible__bu SFEVPKTYVATVHGSVPRNLGRRLREGVELKDGPARVDDFAVVAAVPGKT
MSMEG_3740|M.smegmatis_MC2_155 SFEVPKTYVATVMGTVPRGLGKKLKAGVELDDGPAHVDDFAVVDTVPGKT
MAB_2370|M.abscessus_ATCC_1997 SYEVPKTYIATVAGTIPRGLGRQLRDGVELADGPAKVDEFKFLGTTDGQS
*.**.*:*:*** *::*:.**: * *:** ** . ** * .: : *::
MMAR_2526|M.marinum_M LVRVTLHEGRNRIVRRMLAAVGFPVQALVRIDIGGVSLGEQRPGSVRALR
MUL_3234|M.ulcerans_Agy99 LVRVTLHEGRNRIVRRMLAAVGFPVQALVRIDIGGVSLGEQRPGSVRALR
Mb1738|M.bovis_AF2122/97 LVRVTLHEGRNRIVRRLLAAAGFPVEALVRTDIGAVSLGKQRPGSVRALR
Rv1711|M.tuberculosis_H37Rv LVRVTLHEGRNRIVRRLLAAAGFPVEALVRTDIGAVSLGKQRPGSVRALR
MAV_3065|M.avium_104 LVRVTLHEGRNRIVRRLLAAAGFPVESLVRTDIGPISLGKQRPGSIRALG
MLBr_01370|M.leprae_Br4923 LVRLTLHEGRKRIVRRLLTAAGFPVEMLVRTDIGAVSLGDQRPGCLRALL
Mflv_3495|M.gilvum_PYR-GCK LVRVTLHEGRKHIVRRLLAEVGFPVQNLVRTDIGGVSLGDQRPGSIRALT
Mvan_3276|M.vanbaalenii_PYR-1 LVRVTLHEGRKHIVRRLLAEVGFPVQELVRTDIGTVSLGDQRPGSIRVLT
TH_0200|M.thermoresistible__bu MVRVTLHEGRKRIVRRLLAAVGFPVQDLVRTDIGAVTLGDQRPGSIRALT
MSMEG_3740|M.smegmatis_MC2_155 MVKVTLHEGRNRIVRRLLAAVDFPVQELVRTEIGSVSLGDQRPGSIRALT
MAB_2370|M.abscessus_ATCC_1997 MVQLTLHEGRNRIVRRMMDAAGHPVVELVRTKIGDVTLGNQRPGSLRALG
:*::******::****:: ...** *** .** ::**.****.:*.*
MMAR_2526|M.marinum_M RDEIGQLYKAVGL
MUL_3234|M.ulcerans_Agy99 RDEIGQLYKAVGL
Mb1738|M.bovis_AF2122/97 SNEIGQLYQAVGL
Rv1711|M.tuberculosis_H37Rv SNEIGQLYQAVGL
MAV_3065|M.avium_104 RDEVGRLYQAVGL
MLBr_01370|M.leprae_Br4923 RDEIRQLYKEVGL
Mflv_3495|M.gilvum_PYR-GCK QKEIGELYKAVGL
Mvan_3276|M.vanbaalenii_PYR-1 QKEIGELYKAVGL
TH_0200|M.thermoresistible__bu RKEVGELYKAVGL
MSMEG_3740|M.smegmatis_MC2_155 RKEVGELYQAVGL
MAB_2370|M.abscessus_ATCC_1997 RDEIGSLYKAVGL
.*: **: ***