For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHLNPILGCMPDSRVRATLLGGALGDALGGFVEFDSIEQIRLQFGRHGIAEPPAGQPMLITDDTQMTLFT AEAVIRARRIHDHEPAEIAHRAYLRWLHTQGGHPPADVIDGWLVAVPELHSLRAPGHTCLSALEHTSSAA RERGSRTQRLNNSKGCGAVMRAAPVGFAGADPASIFVMSADIGALTHSHPTGYLSAAALSVIIAEVCAGK TIPVAVETASALLRAEPDHEETYRALRRAVELADTRLTPEQIAARLGGGWVGEEALAIGVYAALAAHDFK DGIRLSVNHSGDSDSTGAIAGNILGAMCGTESLPQDWLDELELREVITTIADDLVGPPGPDWDERYPPR
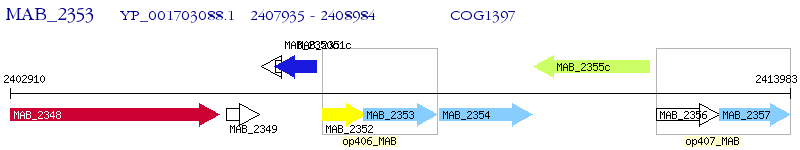
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2353 | - | - | 100% (349) | ADP-ribosylglycohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5181 | - | 2e-12 | 24.47% (331) | ADP-ribosylation/crystallin J1 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1272 | - | 5e-09 | 25.16% (318) | putative ribosylglycohydrolase |
| M. thermoresistible (build 8) | TH_0049 | - | 2e-11 | 27.59% (348) | putative ribosylglycohydrolase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1100 | - | 7e-13 | 24.60% (313) | ADP-ribosylation/crystallin J1 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1272|M.smegmatis_MC2_155 MGSSRIELTAAQRDRAVGVLLGAAAGDALGAGYELNRPLAPDHPVGMIGG
TH_0049|M.thermoresistible__bu ------------------VLLATAAGDALGAGYEFDPPMGDDEPVGMIGG
Mflv_5181|M.gilvum_PYR-GCK --------MTSHDDRIAGVLLGTAAGDALGAPYEFQPPRGPDLDVRMAGG
Mvan_1100|M.vanbaalenii_PYR-1 --------MTARDDRIEGVLLGTAAGDALGAPYEFQPPRGTELDVAMTGG
MAB_2353|M.abscessus_ATCC_1997 -MHLNPILGCMPDSRVRATLLGGALGDALGGFVEFDSIEQIRLQFGRHGI
.**. * *****. *:: . *
MSMEG_1272|M.smegmatis_MC2_155 GL-GPFAPGEWTDDTSMAIAIAEIASTGEDLRKEKPLDYLVERWEWWSRT
TH_0049|M.thermoresistible__bu GL-GPFAPGEWTDDTAMALAIAEIAATGVDLRDDSVQDHIVERWYEWART
Mflv_5181|M.gilvum_PYR-GCK ---GVWEPGEWTDDTAMAIAIAEVAATGADLRDERAQDAILTRWREWSHH
Mvan_1100|M.vanbaalenii_PYR-1 ---GPWTPGEWTDDTAMAVAIAEVAATGADLRDEAAQDAIVARWHEWSRS
MAB_2353|M.abscessus_ATCC_1997 AEPPAGQPMLITDDTQMTLFTAEAVIRARRIHDHEPAEIAHRAYLRWLHT
* **** *:: ** . . ::.. : : * :
MSMEG_1272|M.smegmatis_MC2_155 APDVG--------VQTRTVLSETAARG-LCAQTARAVSEELHRSSGRTA-
TH_0049|M.thermoresistible__bu AKDVG--------NQTSAVLSAVGRRG-ISARAAREESAVWHESAGRSG-
Mflv_5181|M.gilvum_PYR-GCK TKNIG--------VQTSSVLHAASRGAAITAARARDASRAHHARSGRSG-
Mvan_1100|M.vanbaalenii_PYR-1 AKDVG--------IQTRAVLSAAARGGEITAHRARREAGIHHEHTGRSG-
MAB_2353|M.abscessus_ATCC_1997 QGGHPPADVIDGWLVAVPELHSLRAPGHTCLSALEHTSSAARERGSRTQR
. : . * . . : : .*:
MSMEG_1272|M.smegmatis_MC2_155 -----GNGSLMRTSPVALAYLDDEAALVQAARTISELTHFDP----NAGD
TH_0049|M.thermoresistible__bu -----GNGSLMRTAPVALAHLDDEDALADAARAISELTHFDP----EAGD
Mflv_5181|M.gilvum_PYR-GCK -----GNGSLMRTAPVALAYLDDEGAMVEAARAISALTHFDP----EAGD
Mvan_1100|M.vanbaalenii_PYR-1 -----GNGSLMRTAPVALAYLDDEAAMVQGARALSELTHFDP----DAGD
MAB_2353|M.abscessus_ATCC_1997 LNNSKGCGAVMRAAPVGFAGADPASIFVMSA-DIGALTHSHPTGYLSAAA
* *::**::**.:* * :. .* :. *** .* .*.
MSMEG_1272|M.smegmatis_MC2_155 ACVLWCTAIRHTVLTGEIDVRVGLSHIVSERRRLWEARLAEAEASPPSAF
TH_0049|M.thermoresistible__bu ACVMWCLAIRHAVLTGGFDVRIGLDHIDGSRRDLWAARLEEAESSRPEAF
Mflv_5181|M.gilvum_PYR-GCK ACVLWCSAIRHGVLTGDLDVRVGLRHLGDEGRVTWSERLDLAETSRPRDF
Mvan_1100|M.vanbaalenii_PYR-1 ACVLWCSAIRHAVLTGEIDIRIGLGHIDVERRELWATRLEAAERSRPSDF
MAB_2353|M.abscessus_ATCC_1997 LSVIIAEVCAGKTIPVAVETASALLRAEPDHEETYRALRRAVELADTRLT
.*: . . .:. .: .* : . . : .* : .
MSMEG_1272|M.smegmatis_MC2_155 CDNNGWVVAALQAAWSAISTTPVPHEDPRSEVFRADHLRLALEAAVRGGG
TH_0049|M.thermoresistible__bu AGKNGWVVAALQGAWSSIVTTPIPVDDPAAGLFRADHLRLALEAAVRGGG
Mflv_5181|M.gilvum_PYR-GCK P-KNGWVVSALQAAWSAIATTASSEADPNAAG--TGHLPRGLDAAVRAGF
Mvan_1100|M.vanbaalenii_PYR-1 P-KNGWVVQALQAAWSAIATTAVPADDPVAGTFRAGHLRDGLDAAVRAGF
MAB_2353|M.abscessus_ATCC_1997 P---EQIAARLGGGWVGEEALAIGVY----AALAAHDFKDGIRLSVNHSG
:. * ..* . : . : .: .: :*. .
MSMEG_1272|M.smegmatis_MC2_155 DTDTVAAIAGGLLGAAYGASAVPSQWRRLLHGWPGLRTRGLTALADRTVN
TH_0049|M.thermoresistible__bu DTDTVAAIAGGLLGAVHGASAVPAQWRLLLTGWPGLNTRGLVQLATDIVS
Mflv_5181|M.gilvum_PYR-GCK DTDTVAAIAGGLLGAVHGASAVPRAWRELLHGWPGLDAAELAELAHAAAR
Mvan_1100|M.vanbaalenii_PYR-1 DTDTVAAIAGGLLGAAYGASAVPLEWRTLLHGWPGVRARELVKLSGRIAR
MAB_2353|M.abscessus_ATCC_1997 DSDSTGAIAGNILGAMCGTESLPQDW------LDELELREVITTIADDLV
*:*:..****.:*** *:.::* * : :
MSMEG_1272|M.smegmatis_MC2_155 KGRPVEFNQSYAAGRETPEPVRHPHDDGVWIGIAYSLNRLPSDIDAVVSL
TH_0049|M.thermoresistible__bu GGQAESFDQAYGAYPESALAVRHPHDVDIWIGNAAALHRLPDGVDAVVSL
Mflv_5181|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1100|M.vanbaalenii_PYR-1 RGAVDRFDPRYPGY-QVGVMVPHPYDDGVLLAGIGALHELPAGVDAVVSL
MAB_2353|M.abscessus_ATCC_1997 GPPGPDWDERYPPR------------------------------------
MSMEG_1272|M.smegmatis_MC2_155 CRVADGHIPLRVVHLDVPLLDQEG--ANPNLDFVVLDTVRAIEQLRAEGR
TH_0049|M.thermoresistible__bu CRVADEHIPDGATHLDVRLIDRVG--DNANLDFVLLDTVRAVEQLRAENR
Mflv_5181|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1100|M.vanbaalenii_PYR-1 CRVGDEVVRRDIPHVEVRLIDRPGAHDNPHLDFVMLEAVRAVERFRGEGK
MAB_2353|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1272|M.smegmatis_MC2_155 TVFVHSAAAQNRAPAVAALYGARRQGLGIDRALREVCAVLPSAQPNPDLR
TH_0049|M.thermoresistible__bu TVYLHCVHAYSRTPTIAALYGARRQGIGIERALDEVCAVLSGANPNPEFR
Mflv_5181|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1100|M.vanbaalenii_PYR-1 TVLLHCVEAHSRTPSVAALYGARLRGVGTGEALHAVCGALPDASPNPAFR
MAB_2353|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1272|M.smegmatis_MC2_155 AALRRLHVVEAGAR
TH_0049|M.thermoresistible__bu DALRRLHSGD----
Mflv_5181|M.gilvum_PYR-GCK --------------
Mvan_1100|M.vanbaalenii_PYR-1 EALARFDSAHAAAR
MAB_2353|M.abscessus_ATCC_1997 --------------